Neuron
-
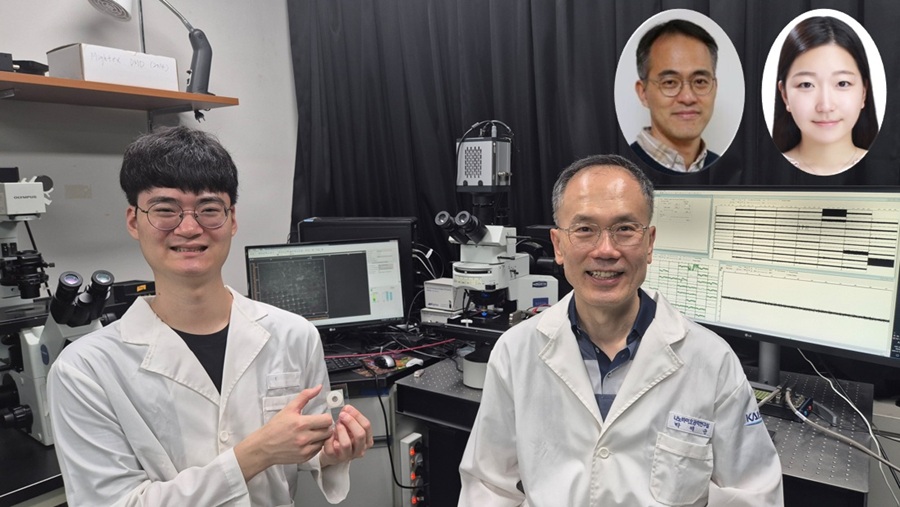 KAIST Successfully Implements 3D Brain-Mimicking Platform with 6x Higher Precision
<(From left) Dr. Dongjo Yoon, Professor Je-Kyun Park from the Department of Bio and Brain Engineering, (upper right) Professor Yoonkey Nam, Dr. Soo Jee Kim>
Existing three-dimensional (3D) neuronal culture technology has limitations in brain research due to the difficulty of precisely replicating the brain's complex multilayered structure and the lack of a platform that can simultaneously analyze both structure and function. A KAIST research team has successfully developed an integrated platform that can implement brain-like layered neuronal structures using 3D printing technology and precisely measure neuronal activity within them.
KAIST (President Kwang Hyung Lee) announced on the 16th of July that a joint research team led by Professors Je-Kyun Park and Yoonkey Nam from the Department of Bio and Brain Engineering has developed an integrated platform capable of fabricating high-resolution 3D multilayer neuronal networks using low-viscosity natural hydrogels with mechanical properties similar to brain tissue, and simultaneously analyzing their structural and functional connectivity.
Conventional bioprinting technology uses high-viscosity bioinks for structural stability, but this limits neuronal proliferation and neurite growth. Conversely, neural cell-friendly low-viscosity hydrogels are difficult to precisely pattern, leading to a fundamental trade-off between structural stability and biological function. The research team completed a sophisticated and stable brain-mimicking platform by combining three key technologies that enable the precise creation of brain structure with dilute gels, accurate alignment between layers, and simultaneous observation of neuronal activity.
The three core technologies are: ▲ 'Capillary Pinning Effect' technology, which enables the dilute gel (hydrogel) to adhere firmly to a stainless steel mesh (micromesh) to prevent it from flowing, thereby reproducing brain structures with six times greater precision (resolution of 500 μm or less) than conventional methods; ▲ the '3D Printing Aligner,' a cylindrical design that ensures the printed layers are precisely stacked without misalignment, guaranteeing the accurate assembly of multilayer structures and stable integration with microelectrode chips; and ▲ 'Dual-mode Analysis System' technology, which simultaneously measures electrical signals from below and observes cell activity with light (calcium imaging) from above, allowing for the simultaneous verification of the functional operation of interlayer connections through multiple methods.
< Figure 1. Platform integrating brain-structure-mimicking neural network model construction and functional measurement technology>
The research team successfully implemented a three-layered mini-brain structure using 3D printing with a fibrin hydrogel, which has elastic properties similar to those of the brain, and experimentally verified the process of actual neural cells transmitting and receiving signals within it.
Cortical neurons were placed in the upper and lower layers, while the middle layer was left empty but designed to allow neurons to penetrate and connect through it. Electrical signals were measured from the lower layer using a microsensor (electrode chip), and cell activity was observed from the upper layer using light (calcium imaging). The results showed that when electrical stimulation was applied, neural cells in both upper and lower layers responded simultaneously. When a synapse-blocking agent (synaptic blocker) was introduced, the response decreased, proving that the neural cells were genuinely connected and transmitting signals.
Professor Je-Kyun Park of KAIST explained, "This research is a joint development achievement of an integrated platform that can simultaneously reproduce the complex multilayered structure and function of brain tissue. Compared to existing technologies where signal measurement was impossible for more than 14 days, this platform maintains a stable microelectrode chip interface for over 27 days, allowing the real-time analysis of structure-function relationships. It can be utilized in various brain research fields such as neurological disease modeling, brain function research, neurotoxicity assessment, and neuroprotective drug screening in the future."
<Figure 2. Integration process of stacked bioprinting technology and microelectrode chip>
The research, in which Dr. Soo Jee Kim and Dr. Dongjo Yoon from KAIST's Department of Bio and Brain Engineering participated as co-first authors, was published online in the international journal 'Biosensors and Bioelectronics' on June 11, 2025.
※Paper: Hybrid biofabrication of multilayered 3D neuronal networks with structural and functional interlayer connectivity
※DOI: https://doi.org/10.1016/j.bios.2025.117688
2025.07.16 View 84
KAIST Successfully Implements 3D Brain-Mimicking Platform with 6x Higher Precision
<(From left) Dr. Dongjo Yoon, Professor Je-Kyun Park from the Department of Bio and Brain Engineering, (upper right) Professor Yoonkey Nam, Dr. Soo Jee Kim>
Existing three-dimensional (3D) neuronal culture technology has limitations in brain research due to the difficulty of precisely replicating the brain's complex multilayered structure and the lack of a platform that can simultaneously analyze both structure and function. A KAIST research team has successfully developed an integrated platform that can implement brain-like layered neuronal structures using 3D printing technology and precisely measure neuronal activity within them.
KAIST (President Kwang Hyung Lee) announced on the 16th of July that a joint research team led by Professors Je-Kyun Park and Yoonkey Nam from the Department of Bio and Brain Engineering has developed an integrated platform capable of fabricating high-resolution 3D multilayer neuronal networks using low-viscosity natural hydrogels with mechanical properties similar to brain tissue, and simultaneously analyzing their structural and functional connectivity.
Conventional bioprinting technology uses high-viscosity bioinks for structural stability, but this limits neuronal proliferation and neurite growth. Conversely, neural cell-friendly low-viscosity hydrogels are difficult to precisely pattern, leading to a fundamental trade-off between structural stability and biological function. The research team completed a sophisticated and stable brain-mimicking platform by combining three key technologies that enable the precise creation of brain structure with dilute gels, accurate alignment between layers, and simultaneous observation of neuronal activity.
The three core technologies are: ▲ 'Capillary Pinning Effect' technology, which enables the dilute gel (hydrogel) to adhere firmly to a stainless steel mesh (micromesh) to prevent it from flowing, thereby reproducing brain structures with six times greater precision (resolution of 500 μm or less) than conventional methods; ▲ the '3D Printing Aligner,' a cylindrical design that ensures the printed layers are precisely stacked without misalignment, guaranteeing the accurate assembly of multilayer structures and stable integration with microelectrode chips; and ▲ 'Dual-mode Analysis System' technology, which simultaneously measures electrical signals from below and observes cell activity with light (calcium imaging) from above, allowing for the simultaneous verification of the functional operation of interlayer connections through multiple methods.
< Figure 1. Platform integrating brain-structure-mimicking neural network model construction and functional measurement technology>
The research team successfully implemented a three-layered mini-brain structure using 3D printing with a fibrin hydrogel, which has elastic properties similar to those of the brain, and experimentally verified the process of actual neural cells transmitting and receiving signals within it.
Cortical neurons were placed in the upper and lower layers, while the middle layer was left empty but designed to allow neurons to penetrate and connect through it. Electrical signals were measured from the lower layer using a microsensor (electrode chip), and cell activity was observed from the upper layer using light (calcium imaging). The results showed that when electrical stimulation was applied, neural cells in both upper and lower layers responded simultaneously. When a synapse-blocking agent (synaptic blocker) was introduced, the response decreased, proving that the neural cells were genuinely connected and transmitting signals.
Professor Je-Kyun Park of KAIST explained, "This research is a joint development achievement of an integrated platform that can simultaneously reproduce the complex multilayered structure and function of brain tissue. Compared to existing technologies where signal measurement was impossible for more than 14 days, this platform maintains a stable microelectrode chip interface for over 27 days, allowing the real-time analysis of structure-function relationships. It can be utilized in various brain research fields such as neurological disease modeling, brain function research, neurotoxicity assessment, and neuroprotective drug screening in the future."
<Figure 2. Integration process of stacked bioprinting technology and microelectrode chip>
The research, in which Dr. Soo Jee Kim and Dr. Dongjo Yoon from KAIST's Department of Bio and Brain Engineering participated as co-first authors, was published online in the international journal 'Biosensors and Bioelectronics' on June 11, 2025.
※Paper: Hybrid biofabrication of multilayered 3D neuronal networks with structural and functional interlayer connectivity
※DOI: https://doi.org/10.1016/j.bios.2025.117688
2025.07.16 View 84 -
 KAIST Shows That the Brain Can Distinguish Glucose: Clues to Treat Obesity and Diabetes
<(From left)Prof. Greg S.B Suh, Dr. Jieun Kim, Dr. Shinhye Kim, Researcher Wongyo Jeong)
“How does our brain distinguish glucose from the many nutrients absorbed in the gut?” Starting with this question, a KAIST research team has demonstrated that the brain can selectively recognize specific nutrients—particularly glucose—beyond simply detecting total calorie content. This study is expected to offer a new paradigm for appetite control and the treatment of metabolic diseases.
On the 9th, KAIST (President Kwang Hyung Lee) announced that Professor Greg S.B. Suh’s team in the Department of Biological Sciences, in collaboration with Professor Young-Gyun Park’s team (BarNeuro), Professor Seung-Hee Lee’s team (Department of Biological Sciences), and the Albert Einstein College of Medicine in New York, had identified the existence of a gut-brain circuit that allows animals in a hungry state to selectively detect and prefer glucose in the gut.
Organisms derive energy from various nutrients including sugars, proteins, and fats. Previous studies have shown that total caloric information in the gut suppresses hunger neurons in the hypothalamus to regulate appetite. However, the existence of a gut-brain circuit that specifically responds to glucose and corresponding brain cells had not been demonstrated until now.
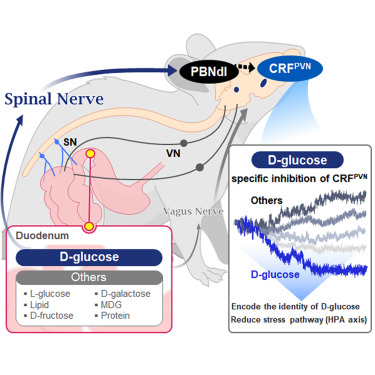
In this study, the team successfully identified a “gut-brain circuit” that senses glucose—essential for brain function—and regulates food intake behavior for required nutrients.
They further proved, for the first time, that this circuit responds within seconds to not only hunger or external stimuli but also to specific caloric nutrients directly introduced into the small intestine, particularly D-glucose, through the activity of “CRF neurons*” in the brain’s hypothalamus.
*CRF neurons: These neurons secrete corticotropin-releasing factor (CRF) in the hypothalamus and are central to the hypothalamic-pituitary-adrenal (HPA) axis, the body’s core physiological system for responding to stress. CRF neurons are known to regulate neuroendocrine balance in response to stress stimuli.
Using optogenetics to precisely track neural activity in real time, the researchers injected various nutrients—D-glucose, L-glucose, amino acids, and fats—directly into the small intestines of mice and observed the results.
They discovered that among the CRF neurons located in the paraventricular nucleus (PVN)* of the hypothalamus, only those specific to D-glucose showed selective responses. These neurons did not respond—or showed inverse reactions—to other sugars or to proteins and fats. This is the first demonstration that single neurons in the brain can guide nutrient-specific responses depending on gut nutrient influx.
*PVN (Paraventricular Nucleus): A key nucleus within the hypothalamus responsible for maintaining bodily homeostasis.
The team also revealed that glucose-sensing signals in the small intestine are transmitted via the spinal cord to the dorsolateral parabrachial nucleus (PBNdl) of the brain, and from there to CRF neurons in the PVN. In contrast, signals for amino acids and fats are transmitted to the brain through the vagus nerve, a different pathway.
In optogenetic inhibition experiments, suppressing CRF neurons in fasting mice eliminated their preference for glucose, proving that this circuit is essential for glucose-specific nutrient preference.
This study was inspired by Professor Suh’s earlier research at NYU using fruit flies, where he identified “DH44 neurons” that selectively detect glucose and sugar in the gut. Based on the hypothesis that hypothalamic neurons in mammals would show similar functional responses to glucose, the current study was launched.
To test this hypothesis, Dr. Jineun Kim (KAIST Ph.D. graduate, now at Caltech) demonstrated during her doctoral research that hungry mice preferred glucose among various intragastrically infused nutrients and that CRF neurons exhibited rapid and specific responses.
Along with Wongyo Jung (KAIST B.S. graduate, now Ph.D. student at Caltech), they modeled and experimentally confirmed the critical role of CRF neurons. Dr. Shinhye Kim, through collaboration, revealed that specific spinal neurons play a key role in conveying intestinal nutrient information to the brain.
Dr. Jineun Kim and Dr. Shinhye Kim said, “This study started from a simple but fundamental question—‘How does the brain distinguish glucose from various nutrients absorbed in the gut?’ We have shown that spinal-based gut-brain circuits play a central role in energy metabolism and homeostasis by transmitting specific gut nutrient signals to the brain.”
Professor Suh added, “By identifying a gut-brain pathway specialized for glucose, this research offers a new therapeutic target for metabolic diseases such as obesity and diabetes. Our future research will explore similar circuits for sensing other essential nutrients like amino acids and fats and their interaction mechanisms.”
Ph.D. student Jineun Kim, Dr. Shinhye Kim, and student Wongyo Jung (co-first authors) contributed to this study, which was published online in the international journal Neuron on June 20, 2025.
※ Paper Title: Encoding the glucose identity by discrete hypothalamic neurons via the gut-brain axis ※ DOI: https://doi.org/10.1016/j.neuron.2025.05.024
This study was supported by the Samsung Science & Technology Foundation, the National Research Foundation of Korea (NRF) Leader Research Program, the POSCO Cheongam Science Fellowship, the Asan Foundation Biomedical Science Scholarship, the Institute for Basic Science (IBS), and the KAIST KAIX program.
2025.07.09 View 268
KAIST Shows That the Brain Can Distinguish Glucose: Clues to Treat Obesity and Diabetes
<(From left)Prof. Greg S.B Suh, Dr. Jieun Kim, Dr. Shinhye Kim, Researcher Wongyo Jeong)
“How does our brain distinguish glucose from the many nutrients absorbed in the gut?” Starting with this question, a KAIST research team has demonstrated that the brain can selectively recognize specific nutrients—particularly glucose—beyond simply detecting total calorie content. This study is expected to offer a new paradigm for appetite control and the treatment of metabolic diseases.
On the 9th, KAIST (President Kwang Hyung Lee) announced that Professor Greg S.B. Suh’s team in the Department of Biological Sciences, in collaboration with Professor Young-Gyun Park’s team (BarNeuro), Professor Seung-Hee Lee’s team (Department of Biological Sciences), and the Albert Einstein College of Medicine in New York, had identified the existence of a gut-brain circuit that allows animals in a hungry state to selectively detect and prefer glucose in the gut.
Organisms derive energy from various nutrients including sugars, proteins, and fats. Previous studies have shown that total caloric information in the gut suppresses hunger neurons in the hypothalamus to regulate appetite. However, the existence of a gut-brain circuit that specifically responds to glucose and corresponding brain cells had not been demonstrated until now.
In this study, the team successfully identified a “gut-brain circuit” that senses glucose—essential for brain function—and regulates food intake behavior for required nutrients.
They further proved, for the first time, that this circuit responds within seconds to not only hunger or external stimuli but also to specific caloric nutrients directly introduced into the small intestine, particularly D-glucose, through the activity of “CRF neurons*” in the brain’s hypothalamus.
*CRF neurons: These neurons secrete corticotropin-releasing factor (CRF) in the hypothalamus and are central to the hypothalamic-pituitary-adrenal (HPA) axis, the body’s core physiological system for responding to stress. CRF neurons are known to regulate neuroendocrine balance in response to stress stimuli.
Using optogenetics to precisely track neural activity in real time, the researchers injected various nutrients—D-glucose, L-glucose, amino acids, and fats—directly into the small intestines of mice and observed the results.
They discovered that among the CRF neurons located in the paraventricular nucleus (PVN)* of the hypothalamus, only those specific to D-glucose showed selective responses. These neurons did not respond—or showed inverse reactions—to other sugars or to proteins and fats. This is the first demonstration that single neurons in the brain can guide nutrient-specific responses depending on gut nutrient influx.
*PVN (Paraventricular Nucleus): A key nucleus within the hypothalamus responsible for maintaining bodily homeostasis.
The team also revealed that glucose-sensing signals in the small intestine are transmitted via the spinal cord to the dorsolateral parabrachial nucleus (PBNdl) of the brain, and from there to CRF neurons in the PVN. In contrast, signals for amino acids and fats are transmitted to the brain through the vagus nerve, a different pathway.
In optogenetic inhibition experiments, suppressing CRF neurons in fasting mice eliminated their preference for glucose, proving that this circuit is essential for glucose-specific nutrient preference.
This study was inspired by Professor Suh’s earlier research at NYU using fruit flies, where he identified “DH44 neurons” that selectively detect glucose and sugar in the gut. Based on the hypothesis that hypothalamic neurons in mammals would show similar functional responses to glucose, the current study was launched.
To test this hypothesis, Dr. Jineun Kim (KAIST Ph.D. graduate, now at Caltech) demonstrated during her doctoral research that hungry mice preferred glucose among various intragastrically infused nutrients and that CRF neurons exhibited rapid and specific responses.
Along with Wongyo Jung (KAIST B.S. graduate, now Ph.D. student at Caltech), they modeled and experimentally confirmed the critical role of CRF neurons. Dr. Shinhye Kim, through collaboration, revealed that specific spinal neurons play a key role in conveying intestinal nutrient information to the brain.
Dr. Jineun Kim and Dr. Shinhye Kim said, “This study started from a simple but fundamental question—‘How does the brain distinguish glucose from various nutrients absorbed in the gut?’ We have shown that spinal-based gut-brain circuits play a central role in energy metabolism and homeostasis by transmitting specific gut nutrient signals to the brain.”
Professor Suh added, “By identifying a gut-brain pathway specialized for glucose, this research offers a new therapeutic target for metabolic diseases such as obesity and diabetes. Our future research will explore similar circuits for sensing other essential nutrients like amino acids and fats and their interaction mechanisms.”
Ph.D. student Jineun Kim, Dr. Shinhye Kim, and student Wongyo Jung (co-first authors) contributed to this study, which was published online in the international journal Neuron on June 20, 2025.
※ Paper Title: Encoding the glucose identity by discrete hypothalamic neurons via the gut-brain axis ※ DOI: https://doi.org/10.1016/j.neuron.2025.05.024
This study was supported by the Samsung Science & Technology Foundation, the National Research Foundation of Korea (NRF) Leader Research Program, the POSCO Cheongam Science Fellowship, the Asan Foundation Biomedical Science Scholarship, the Institute for Basic Science (IBS), and the KAIST KAIX program.
2025.07.09 View 268 -
 KAIST Presents Game-Changing Technology for Intractable Brain Disease Treatment Using Micro OLEDs
<(From left)Professor Kyung Cheol Choi, Hyunjoo J. Lee, Somin Lee from the School of Electrical Engineering>
Optogenetics is a technique that controls neural activity by stimulating neurons expressing light-sensitive proteins with specific wavelengths of light. It has opened new possibilities for identifying causes of brain disorders and developing treatments for intractable neurological diseases. Because this technology requires precise stimulation inside the human brain with minimal damage to soft brain tissue, it must be integrated into a neural probe—a medical device implanted in the brain. KAIST researchers have now proposed a new paradigm for neural probes by integrating micro OLEDs into thin, flexible, implantable medical devices.
KAIST (President Kwang Hyung Lee) announced on the 6th of July that Professor Kyung Cheol Choi and researcher Hyunjoo J. Lee from the School of Electrical Engineering have jointly succeeded in developing an optogenetic neural probe integrated with flexible micro OLEDs.
Optical fibers have been used for decades in optogenetic research to deliver light to deep brain regions from external light sources. Recently, research has focused on flexible optical fibers and ultra-miniaturized neural probes that integrate light sources for single-neuron stimulation.
The research team focused on micro OLEDs due to their high spatial resolution and flexibility, which allow for precise light delivery to small areas of neurons. This enables detailed brain circuit analysis while minimizing side effects and avoiding restrictions on animal movement. Moreover, micro OLEDs offer precise control of light wavelengths and support multi-site stimulation, making them suitable for studying complex brain functions.
However, the device's electrical properties degrade easily in the presence of moisture or water, which limited their use as implantable bioelectronics. Furthermore, optimizing the high-resolution integration process on thin, flexible probes remained a challenge.
To address this, the team enhanced the operational reliability of OLEDs in moist, oxygen-rich environments and minimized tissue damage during implantation. They patterned an ultrathin, flexible encapsulation layer* composed of aluminum oxide and parylene-C (Al₂O₃/parylene-C) at widths of 260–600 micrometers (μm) to maintain biocompatibility.
*Encapsulation layer: A barrier that completely blocks oxygen and water molecules from the external environment, ensuring the longevity and reliability of the device.
When integrating the high-resolution micro OLEDs, the researchers also used parylene-C, the same biocompatible material as the encapsulation layer, to maintain flexibility and safety. To eliminate electrical interference between adjacent OLED pixels and spatially separate them, they introduced a pixel define layer (PDL), enabling the independent operation of eight micro OLEDs.
Furthermore, they precisely controlled the residual stress and thickness in the multilayer film structure of the device, ensuring its flexibility even in biological environments. This optimization allowed for probe insertion without bending or external shuttles or needles, minimizing mechanical stress during implantation.
Advanced Functional Materials-Conceptual diagram of a flexible neural probe for integrated optogenetics (Micro-OLED)>
As a result, the team developed a flexible neural probe with integrated micro OLEDs capable of emitting more than one milliwatt per square millimeter (mW/mm²) at 470 nanometers (nm), the optimal wavelength for activating channelrhodopsin-2. This is a significantly high light output for optogenetics and biomedical stimulation applications.
The ultrathin flexible encapsulation layer exhibited a low water vapor transmission rate of 2.66×10⁻⁵ g/m²/day, allowing the device to maintain functionality for over 10 years. The parylene-C-based barrier also demonstrated excellent performance in biological environments, successfully enabling the independent operation of the integrated OLEDs without electrical interference or bending issues.
Dr. Somin Lee, the lead author from Professor Choi’s lab, stated, “We focused on fine-tuning the integration process of highly flexible, high-resolution micro OLEDs onto thin flexible probes, enhancing their biocompatibility and application potential. This is the first reported development of such flexible OLEDs in a probe format and presents a new paradigm for using flexible OLEDs as implantable medical devices for monitoring and therapy.”
This study, with Dr. Somin Lee as the first author, was published online on March 26 in Advanced Functional Materials (IF 18.5), a leading international journal in the field of nanotechnology, and was selected as the cover article for the upcoming July issue.
※ Title: Advanced Micro-OLED Integration on Thin and Flexible Polymer Neural Probes for Targeted Optogenetic Stimulation ※ DOI: https://doi.org/10.1002/adfm.202420758
The research was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Electronic Medicine Technology Development Program (Project title: Development of Core Source Technologies and In Vivo Validation for Brain Cognition and Emotion-Enhancing Light-Stimulating Electronic Medicine).
2025.07.07 View 238
KAIST Presents Game-Changing Technology for Intractable Brain Disease Treatment Using Micro OLEDs
<(From left)Professor Kyung Cheol Choi, Hyunjoo J. Lee, Somin Lee from the School of Electrical Engineering>
Optogenetics is a technique that controls neural activity by stimulating neurons expressing light-sensitive proteins with specific wavelengths of light. It has opened new possibilities for identifying causes of brain disorders and developing treatments for intractable neurological diseases. Because this technology requires precise stimulation inside the human brain with minimal damage to soft brain tissue, it must be integrated into a neural probe—a medical device implanted in the brain. KAIST researchers have now proposed a new paradigm for neural probes by integrating micro OLEDs into thin, flexible, implantable medical devices.
KAIST (President Kwang Hyung Lee) announced on the 6th of July that Professor Kyung Cheol Choi and researcher Hyunjoo J. Lee from the School of Electrical Engineering have jointly succeeded in developing an optogenetic neural probe integrated with flexible micro OLEDs.
Optical fibers have been used for decades in optogenetic research to deliver light to deep brain regions from external light sources. Recently, research has focused on flexible optical fibers and ultra-miniaturized neural probes that integrate light sources for single-neuron stimulation.
The research team focused on micro OLEDs due to their high spatial resolution and flexibility, which allow for precise light delivery to small areas of neurons. This enables detailed brain circuit analysis while minimizing side effects and avoiding restrictions on animal movement. Moreover, micro OLEDs offer precise control of light wavelengths and support multi-site stimulation, making them suitable for studying complex brain functions.
However, the device's electrical properties degrade easily in the presence of moisture or water, which limited their use as implantable bioelectronics. Furthermore, optimizing the high-resolution integration process on thin, flexible probes remained a challenge.
To address this, the team enhanced the operational reliability of OLEDs in moist, oxygen-rich environments and minimized tissue damage during implantation. They patterned an ultrathin, flexible encapsulation layer* composed of aluminum oxide and parylene-C (Al₂O₃/parylene-C) at widths of 260–600 micrometers (μm) to maintain biocompatibility.
*Encapsulation layer: A barrier that completely blocks oxygen and water molecules from the external environment, ensuring the longevity and reliability of the device.
When integrating the high-resolution micro OLEDs, the researchers also used parylene-C, the same biocompatible material as the encapsulation layer, to maintain flexibility and safety. To eliminate electrical interference between adjacent OLED pixels and spatially separate them, they introduced a pixel define layer (PDL), enabling the independent operation of eight micro OLEDs.
Furthermore, they precisely controlled the residual stress and thickness in the multilayer film structure of the device, ensuring its flexibility even in biological environments. This optimization allowed for probe insertion without bending or external shuttles or needles, minimizing mechanical stress during implantation.
Advanced Functional Materials-Conceptual diagram of a flexible neural probe for integrated optogenetics (Micro-OLED)>
As a result, the team developed a flexible neural probe with integrated micro OLEDs capable of emitting more than one milliwatt per square millimeter (mW/mm²) at 470 nanometers (nm), the optimal wavelength for activating channelrhodopsin-2. This is a significantly high light output for optogenetics and biomedical stimulation applications.
The ultrathin flexible encapsulation layer exhibited a low water vapor transmission rate of 2.66×10⁻⁵ g/m²/day, allowing the device to maintain functionality for over 10 years. The parylene-C-based barrier also demonstrated excellent performance in biological environments, successfully enabling the independent operation of the integrated OLEDs without electrical interference or bending issues.
Dr. Somin Lee, the lead author from Professor Choi’s lab, stated, “We focused on fine-tuning the integration process of highly flexible, high-resolution micro OLEDs onto thin flexible probes, enhancing their biocompatibility and application potential. This is the first reported development of such flexible OLEDs in a probe format and presents a new paradigm for using flexible OLEDs as implantable medical devices for monitoring and therapy.”
This study, with Dr. Somin Lee as the first author, was published online on March 26 in Advanced Functional Materials (IF 18.5), a leading international journal in the field of nanotechnology, and was selected as the cover article for the upcoming July issue.
※ Title: Advanced Micro-OLED Integration on Thin and Flexible Polymer Neural Probes for Targeted Optogenetic Stimulation ※ DOI: https://doi.org/10.1002/adfm.202420758
The research was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Electronic Medicine Technology Development Program (Project title: Development of Core Source Technologies and In Vivo Validation for Brain Cognition and Emotion-Enhancing Light-Stimulating Electronic Medicine).
2025.07.07 View 238 -
 Editing Parkinson's Disease – KAIST Makes World's First Discovery of an Inflammatory RNA Editing Enzyme through Co-work with UCL Researchers
< Professor Minee Choi of the Department of Brain and Cognitive Sciences (top left). Professor Sonia Gandhi (top right) and Professor Klenerman of the University College London (bottom right) >
Parkinson's disease (PD) is a neurodegenerative disorder in which the α-synuclein protein abnormally aggregates within brain cells, causing neuronal damage. Through international collaboration, researchers at KAIST have revealed that RNA editing plays a crucial role in regulating neuroinflammation, a key pathology of Parkinson's disease.
KAIST (represented by President Kwang-Hyung Lee) announced on the 27th of April that a research team led by Professor Minee L. Choi from the Department of Brain and Cognitive Sciences, in collaboration with University College London (UCL) and the Francis Crick Institute, discovered that the RNA editing enzyme ADAR1 plays an important role in controlling immune responses in astrocytes, glial cells that trigger protective reactions in the brain, and demonstrated that this mechanism is critically involved in the progression of Parkinson’s disease.
Professor Choi's research team created a co-culture model composed of astrocytes and neurons derived from stem cells originating from Parkinson's disease patients, in order to study the inflammatory responses of brain immune cells. They then treated the model with α-synuclein aggregates, which are known to cause Parkinson’s disease, and analyzed how the immune cells' inflammatory responses changed.
< Figure 1. Schematic diagram of the inflammatory RNA editing model in Parkinson's disease >
As a result, it was found that early pathological forms of α-synuclein, known as oligomers, activated the Toll-like receptor pathway, which acts as a danger sensor in astrocytes, as well as the interferon response pathway, an immune signaling network that combats viruses and pathogens. During this process, the RNA editing enzyme ADAR1 was expressed and transformed into an isoform with an altered protein structure and function.
Notably, the RNA editing activity of ADAR1, which normally functions to regulate immune responses during viral infections by converting adenosine (A) to inosine (I) through a process known as A-to-I RNA editing, was found to be abnormally focused on genes that cause inflammation rather than operating under normal conditions. This phenomenon was observed not only in the patient-derived neuron models but also in postmortem brain tissues from actual Parkinson’s disease patients.
< Figure 2. Experimental design and inflammatory response induction in astrocytes following treatment with α-synuclein oligomers (abnormally folded protein fragments) >
This directly proves that the dysregulation of RNA editing induces chronic inflammatory responses in astrocytes, ultimately leading to neuronal toxicity and pathological progression.
This study is significant in that it newly identified the regulation of RNA editing within astrocytes as a key mechanism behind neuroinflammatory responses. In particular, it suggests that ADAR1 could serve as a novel genetic target for the treatment of Parkinson’s disease.
It is also noteworthy that the study reflected actual pathological characteristics of patients by utilizing patient-specific induced pluripotent stem cell-based precision models for brain diseases.
Professor Minee L. Choi stated, “This study demonstrates that the regulator of inflammation caused by protein aggregation operates at the new layer of RNA editing, offering a completely different therapeutic strategy from existing approaches to Parkinson's disease treatment." She further emphasized, “RNA editing technology could become an important turning point in the development of therapeutics for neuroinflammation.”
< Figure 3. When treated with α-synuclein oligomers, the causative agent of Parkinson's disease, A-to-I RNA editing is induced to change genetic information by ADAR in patient-derived stem cell-differentiated glial cells, confirming that α-synuclein is likely to be associated with the progression of Parkinson's disease through RNA editing >
This study was published in Science Advances on April 11, with Professor Choi listed as a co-first author.
Paper Title: Astrocytic RNA editing regulates the host immune response to alpha-synuclein, Science Advances Vol.11, Issue 15. (DOI:10.1126/sciadv.adp8504)
Lead Authors: Karishma D’Sa (UCL, Co-First Author), Minee L. Choi (KAIST, Co-First Author), Mina Ryten (UCL, Corresponding Author), Sonia Gandhi (Francis Crick Institute, University of Cambridge, Corresponding Author)
This research was supported by the Brain Research Program and the Excellent Young Researcher Program of the National Research Foundation of Korea, as well as KAIST’s Daekyo Cognitive Enhancement Program.
2025.05.02 View 4726
Editing Parkinson's Disease – KAIST Makes World's First Discovery of an Inflammatory RNA Editing Enzyme through Co-work with UCL Researchers
< Professor Minee Choi of the Department of Brain and Cognitive Sciences (top left). Professor Sonia Gandhi (top right) and Professor Klenerman of the University College London (bottom right) >
Parkinson's disease (PD) is a neurodegenerative disorder in which the α-synuclein protein abnormally aggregates within brain cells, causing neuronal damage. Through international collaboration, researchers at KAIST have revealed that RNA editing plays a crucial role in regulating neuroinflammation, a key pathology of Parkinson's disease.
KAIST (represented by President Kwang-Hyung Lee) announced on the 27th of April that a research team led by Professor Minee L. Choi from the Department of Brain and Cognitive Sciences, in collaboration with University College London (UCL) and the Francis Crick Institute, discovered that the RNA editing enzyme ADAR1 plays an important role in controlling immune responses in astrocytes, glial cells that trigger protective reactions in the brain, and demonstrated that this mechanism is critically involved in the progression of Parkinson’s disease.
Professor Choi's research team created a co-culture model composed of astrocytes and neurons derived from stem cells originating from Parkinson's disease patients, in order to study the inflammatory responses of brain immune cells. They then treated the model with α-synuclein aggregates, which are known to cause Parkinson’s disease, and analyzed how the immune cells' inflammatory responses changed.
< Figure 1. Schematic diagram of the inflammatory RNA editing model in Parkinson's disease >
As a result, it was found that early pathological forms of α-synuclein, known as oligomers, activated the Toll-like receptor pathway, which acts as a danger sensor in astrocytes, as well as the interferon response pathway, an immune signaling network that combats viruses and pathogens. During this process, the RNA editing enzyme ADAR1 was expressed and transformed into an isoform with an altered protein structure and function.
Notably, the RNA editing activity of ADAR1, which normally functions to regulate immune responses during viral infections by converting adenosine (A) to inosine (I) through a process known as A-to-I RNA editing, was found to be abnormally focused on genes that cause inflammation rather than operating under normal conditions. This phenomenon was observed not only in the patient-derived neuron models but also in postmortem brain tissues from actual Parkinson’s disease patients.
< Figure 2. Experimental design and inflammatory response induction in astrocytes following treatment with α-synuclein oligomers (abnormally folded protein fragments) >
This directly proves that the dysregulation of RNA editing induces chronic inflammatory responses in astrocytes, ultimately leading to neuronal toxicity and pathological progression.
This study is significant in that it newly identified the regulation of RNA editing within astrocytes as a key mechanism behind neuroinflammatory responses. In particular, it suggests that ADAR1 could serve as a novel genetic target for the treatment of Parkinson’s disease.
It is also noteworthy that the study reflected actual pathological characteristics of patients by utilizing patient-specific induced pluripotent stem cell-based precision models for brain diseases.
Professor Minee L. Choi stated, “This study demonstrates that the regulator of inflammation caused by protein aggregation operates at the new layer of RNA editing, offering a completely different therapeutic strategy from existing approaches to Parkinson's disease treatment." She further emphasized, “RNA editing technology could become an important turning point in the development of therapeutics for neuroinflammation.”
< Figure 3. When treated with α-synuclein oligomers, the causative agent of Parkinson's disease, A-to-I RNA editing is induced to change genetic information by ADAR in patient-derived stem cell-differentiated glial cells, confirming that α-synuclein is likely to be associated with the progression of Parkinson's disease through RNA editing >
This study was published in Science Advances on April 11, with Professor Choi listed as a co-first author.
Paper Title: Astrocytic RNA editing regulates the host immune response to alpha-synuclein, Science Advances Vol.11, Issue 15. (DOI:10.1126/sciadv.adp8504)
Lead Authors: Karishma D’Sa (UCL, Co-First Author), Minee L. Choi (KAIST, Co-First Author), Mina Ryten (UCL, Corresponding Author), Sonia Gandhi (Francis Crick Institute, University of Cambridge, Corresponding Author)
This research was supported by the Brain Research Program and the Excellent Young Researcher Program of the National Research Foundation of Korea, as well as KAIST’s Daekyo Cognitive Enhancement Program.
2025.05.02 View 4726 -
 KAIST Research Team Breaks Down Musical Instincts with AI
Music, often referred to as the universal language, is known to be a common component in all cultures. Then, could ‘musical instinct’ be something that is shared to some degree despite the extensive environmental differences amongst cultures?
On January 16, a KAIST research team led by Professor Hawoong Jung from the Department of Physics announced to have identified the principle by which musical instincts emerge from the human brain without special learning using an artificial neural network model.
Previously, many researchers have attempted to identify the similarities and differences between the music that exist in various different cultures, and tried to understand the origin of the universality. A paper published in Science in 2019 had revealed that music is produced in all ethnographically distinct cultures, and that similar forms of beats and tunes are used. Neuroscientist have also previously found out that a specific part of the human brain, namely the auditory cortex, is responsible for processing musical information.
Professor Jung’s team used an artificial neural network model to show that cognitive functions for music forms spontaneously as a result of processing auditory information received from nature, without being taught music. The research team utilized AudioSet, a large-scale collection of sound data provided by Google, and taught the artificial neural network to learn the various sounds. Interestingly, the research team discovered that certain neurons within the network model would respond selectively to music. In other words, they observed the spontaneous generation of neurons that reacted minimally to various other sounds like those of animals, nature, or machines, but showed high levels of response to various forms of music including both instrumental and vocal.
The neurons in the artificial neural network model showed similar reactive behaviours to those in the auditory cortex of a real brain. For example, artificial neurons responded less to the sound of music that was cropped into short intervals and were rearranged. This indicates that the spontaneously-generated music-selective neurons encode the temporal structure of music. This property was not limited to a specific genre of music, but emerged across 25 different genres including classic, pop, rock, jazz, and electronic.
< Figure 1. Illustration of the musicality of the brain and artificial neural network (created with DALL·E3 AI based on the paper content) >
Furthermore, suppressing the activity of the music-selective neurons was found to greatly impede the cognitive accuracy for other natural sounds. That is to say, the neural function that processes musical information helps process other sounds, and that ‘musical ability’ may be an instinct formed as a result of an evolutionary adaptation acquired to better process sounds from nature.
Professor Hawoong Jung, who advised the research, said, “The results of our study imply that evolutionary pressure has contributed to forming the universal basis for processing musical information in various cultures.” As for the significance of the research, he explained, “We look forward for this artificially built model with human-like musicality to become an original model for various applications including AI music generation, musical therapy, and for research in musical cognition.” He also commented on its limitations, adding, “This research however does not take into consideration the developmental process that follows the learning of music, and it must be noted that this is a study on the foundation of processing musical information in early development.”
< Figure 2. The artificial neural network that learned to recognize non-musical natural sounds in the cyber space distinguishes between music and non-music. >
This research, conducted by first author Dr. Gwangsu Kim of the KAIST Department of Physics (current affiliation: MIT Department of Brain and Cognitive Sciences) and Dr. Dong-Kyum Kim (current affiliation: IBS) was published in Nature Communications under the title, “Spontaneous emergence of rudimentary music detectors in deep neural networks”.
This research was supported by the National Research Foundation of Korea.
2024.01.23 View 8708
KAIST Research Team Breaks Down Musical Instincts with AI
Music, often referred to as the universal language, is known to be a common component in all cultures. Then, could ‘musical instinct’ be something that is shared to some degree despite the extensive environmental differences amongst cultures?
On January 16, a KAIST research team led by Professor Hawoong Jung from the Department of Physics announced to have identified the principle by which musical instincts emerge from the human brain without special learning using an artificial neural network model.
Previously, many researchers have attempted to identify the similarities and differences between the music that exist in various different cultures, and tried to understand the origin of the universality. A paper published in Science in 2019 had revealed that music is produced in all ethnographically distinct cultures, and that similar forms of beats and tunes are used. Neuroscientist have also previously found out that a specific part of the human brain, namely the auditory cortex, is responsible for processing musical information.
Professor Jung’s team used an artificial neural network model to show that cognitive functions for music forms spontaneously as a result of processing auditory information received from nature, without being taught music. The research team utilized AudioSet, a large-scale collection of sound data provided by Google, and taught the artificial neural network to learn the various sounds. Interestingly, the research team discovered that certain neurons within the network model would respond selectively to music. In other words, they observed the spontaneous generation of neurons that reacted minimally to various other sounds like those of animals, nature, or machines, but showed high levels of response to various forms of music including both instrumental and vocal.
The neurons in the artificial neural network model showed similar reactive behaviours to those in the auditory cortex of a real brain. For example, artificial neurons responded less to the sound of music that was cropped into short intervals and were rearranged. This indicates that the spontaneously-generated music-selective neurons encode the temporal structure of music. This property was not limited to a specific genre of music, but emerged across 25 different genres including classic, pop, rock, jazz, and electronic.
< Figure 1. Illustration of the musicality of the brain and artificial neural network (created with DALL·E3 AI based on the paper content) >
Furthermore, suppressing the activity of the music-selective neurons was found to greatly impede the cognitive accuracy for other natural sounds. That is to say, the neural function that processes musical information helps process other sounds, and that ‘musical ability’ may be an instinct formed as a result of an evolutionary adaptation acquired to better process sounds from nature.
Professor Hawoong Jung, who advised the research, said, “The results of our study imply that evolutionary pressure has contributed to forming the universal basis for processing musical information in various cultures.” As for the significance of the research, he explained, “We look forward for this artificially built model with human-like musicality to become an original model for various applications including AI music generation, musical therapy, and for research in musical cognition.” He also commented on its limitations, adding, “This research however does not take into consideration the developmental process that follows the learning of music, and it must be noted that this is a study on the foundation of processing musical information in early development.”
< Figure 2. The artificial neural network that learned to recognize non-musical natural sounds in the cyber space distinguishes between music and non-music. >
This research, conducted by first author Dr. Gwangsu Kim of the KAIST Department of Physics (current affiliation: MIT Department of Brain and Cognitive Sciences) and Dr. Dong-Kyum Kim (current affiliation: IBS) was published in Nature Communications under the title, “Spontaneous emergence of rudimentary music detectors in deep neural networks”.
This research was supported by the National Research Foundation of Korea.
2024.01.23 View 8708 -
 A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 8179
A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time.
On January 9, a joint research team led by Professor Won Do Heo from the KAIST Department of Biological Sciences, Professor Hyung-Bae Kwon from Johns Hopkins School of Medicine, and Professor Sangkyu Lee from the Institute for Basic Science (IBS) revealed that they have developed the world’s first technique to allow a real-time observation of synapse formation, extinction, and alterations.
Professor Heo’s team conjugated dimerization-dependent fluorescent proteins (ddFP) to synapses in order to observe the process in which synapses create connections between neurons in real time. The team named this technique SynapShot, by combining the words ‘synapse’ and snapshot’, and successfully tracked and observed the live formation and extinction processes of synapses as well as their dynamic changes.
< Figure 1. To observe dynamically changing synapses, dimerization-dependent fluorescent protein (ddFP) was expressed to observe flourescent signals upon synapse formation as ddFP enables fluorescence detection through reversible binding to pre- and postsynaptic terminals. >
Through a joint research project, the teams led by Professor Heo and Professor Sangkyu Lee at IBS together designed a SynapShot with green and red fluorescence, and were able to easily distinguish the synapse connecting two different neurons. Additionally, by combining an optogenetic technique that can control the function of a molecule using light, the team was able to observe the changes in the synapses while simultaneously inducing certain functions of the neurons using light.
Through more joint research with the team led by Professor Hyung-Bae Kwon at the Johns Hopkins School of Medicine, Professor Heo’s team induced several situations on live mice, including visual discrimination training, exercise, and anaesthesia, and used SynapShot to observe the changes in the synapses during each situation in real time. The observations revealed that each synapse could change fairly quickly and dynamically. This was the first-ever case in which the changes in synapses were observed in a live mammal.
< Figure 2. Microscopic photos observed through changes of the flourescence of the synapse sensor (SynapShot) by cultivating the neurons of an experimental rat and expressing the SynapShot. The changes in the synapse that is created when the pre- and post-synaptic terminals come into contact and the synapse that disappears after a certain period of time are measured by the fluorescence of the SynapShot. >
Professor Heo said, “Our group developed SynapShot through a collaboration with domestic and international research teams, and have opened up the possibility for first-hand live observations of the quick and dynamic changes of synapses, which was previously difficult to do. We expect this technique to revolutionize research methodology in the neurological field, and play an important role in brightening the future of brain science.”
This research, conducted by co-first authors Seungkyu Son (Ph.D. candidate), Jinsu Lee (Ph.D. candidate) and Dr. Kanghoon Jung from Johns Hopkins, was published in the online edition of Nature Methods on January 8 under the title “Real-time visualization of structural dynamics of synapses in live cells in vivo”, and will be printed in the February volume.
< Figure 3. Simultaneous use of green-SynapShot and red-SynapShot to distinguish and observe synapses with one post-terminal and different pre-terminals. >
< Figure 4. Dimer-dependent fluorescent protein (ddFP) exists as a green fluorescent protein as well as a red fluorescent protein, and can be applied together with blue light-activated optogenetic technology. After activating Tropomyosin receptor kinase B (TrkB) by blue light using optogenetic technology, the strengthening of synaptic connections through signals of brain-derived neurotrophic factor is observed using red-SynapShot. >
< Figure 5. Micrographs showing real-time changing synapses in the visual cortex of mice trained through visual training using in vivo imaging techniques such as two-photon microscopy as well as at the cellular level. >
This research was supported by Mid-Sized Research Funds and the Singularity Project from KAIST, and by IBS.
2024.01.18 View 8179 -
 Professor Shinhyun Choi’s team, selected for Nature Communications Editors’ highlight
[ From left, Ph.D. candidates See-On Park and Hakcheon Jeong, along with Master's student Jong-Yong Park and Professor Shinhyun Choi ]
See-On Park, Hakcheon Jeong, Jong-Yong Park - a team of researchers under the leadership of Professor Shinhyun Choi of the School of Electrical Engineering, developed a highly reliable variable resistor (memristor) array that simulates the behavior of neurons using a metal oxide layer with an oxygen concentration gradient, and published their work in Nature Communications. The study was selected as the Nature Communications' Editor's highlight, and as the featured article posted on the main page of the journal's website.
Link : https://www.nature.com/ncomms/
[ Figure 1. The featured image on the main page of the Nature Communications' website introducing the research by Professor Choi's team on the memristor for artificial neurons ]
Thesis title: Experimental demonstration of highly reliable dynamic memristor for artificial neuron and neuromorphic computing.
( https://doi.org/10.1038/s41467-022-30539-6 )
At KAIST, their research was introduced on the 2022 Fall issue of Breakthroughs, the biannual newsletter published by KAIST College of Engineering.
This research was conducted with the support from the Samsung Research Funding & Incubation Center of Samsung Electronics.
2022.11.01 View 11066
Professor Shinhyun Choi’s team, selected for Nature Communications Editors’ highlight
[ From left, Ph.D. candidates See-On Park and Hakcheon Jeong, along with Master's student Jong-Yong Park and Professor Shinhyun Choi ]
See-On Park, Hakcheon Jeong, Jong-Yong Park - a team of researchers under the leadership of Professor Shinhyun Choi of the School of Electrical Engineering, developed a highly reliable variable resistor (memristor) array that simulates the behavior of neurons using a metal oxide layer with an oxygen concentration gradient, and published their work in Nature Communications. The study was selected as the Nature Communications' Editor's highlight, and as the featured article posted on the main page of the journal's website.
Link : https://www.nature.com/ncomms/
[ Figure 1. The featured image on the main page of the Nature Communications' website introducing the research by Professor Choi's team on the memristor for artificial neurons ]
Thesis title: Experimental demonstration of highly reliable dynamic memristor for artificial neuron and neuromorphic computing.
( https://doi.org/10.1038/s41467-022-30539-6 )
At KAIST, their research was introduced on the 2022 Fall issue of Breakthroughs, the biannual newsletter published by KAIST College of Engineering.
This research was conducted with the support from the Samsung Research Funding & Incubation Center of Samsung Electronics.
2022.11.01 View 11066 -
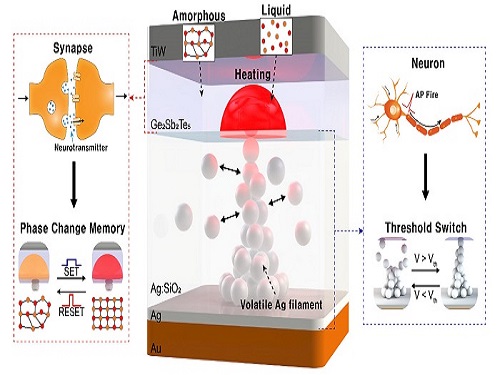 Neuromorphic Memory Device Simulates Neurons and Synapses
Simultaneous emulation of neuronal and synaptic properties promotes the development of brain-like artificial intelligence
Researchers have reported a nano-sized neuromorphic memory device that emulates neurons and synapses simultaneously in a unit cell, another step toward completing the goal of neuromorphic computing designed to rigorously mimic the human brain with semiconductor devices.
Neuromorphic computing aims to realize artificial intelligence (AI) by mimicking the mechanisms of neurons and synapses that make up the human brain. Inspired by the cognitive functions of the human brain that current computers cannot provide, neuromorphic devices have been widely investigated. However, current Complementary Metal-Oxide Semiconductor (CMOS)-based neuromorphic circuits simply connect artificial neurons and synapses without synergistic interactions, and the concomitant implementation of neurons and synapses still remains a challenge. To address these issues, a research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering implemented the biological working mechanisms of humans by introducing the neuron-synapse interactions in a single memory cell, rather than the conventional approach of electrically connecting artificial neuronal and synaptic devices.
Similar to commercial graphics cards, the artificial synaptic devices previously studied often used to accelerate parallel computations, which shows clear differences from the operational mechanisms of the human brain. The research team implemented the synergistic interactions between neurons and synapses in the neuromorphic memory device, emulating the mechanisms of the biological neural network. In addition, the developed neuromorphic device can replace complex CMOS neuron circuits with a single device, providing high scalability and cost efficiency.
The human brain consists of a complex network of 100 billion neurons and 100 trillion synapses. The functions and structures of neurons and synapses can flexibly change according to the external stimuli, adapting to the surrounding environment. The research team developed a neuromorphic device in which short-term and long-term memories coexist using volatile and non-volatile memory devices that mimic the characteristics of neurons and synapses, respectively. A threshold switch device is used as volatile memory and phase-change memory is used as a non-volatile device. Two thin-film devices are integrated without intermediate electrodes, implementing the functional adaptability of neurons and synapses in the neuromorphic memory.
Professor Keon Jae Lee explained, "Neurons and synapses interact with each other to establish cognitive functions such as memory and learning, so simulating both is an essential element for brain-inspired artificial intelligence. The developed neuromorphic memory device also mimics the retraining effect that allows quick learning of the forgotten information by implementing a positive feedback effect between neurons and synapses.”
This result entitled “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse” was published in the May 19, 2022 issue of Nature Communications.
-Publication:Sang Hyun Sung, Tae Jin Kim, Hyera Shin, Tae Hong Im, and Keon Jae Lee (2022) “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse,” Nature Communications May 19, 2022 (DOI: 10.1038/s41467-022-30432-2)
-Profile:Professor Keon Jae Leehttp://fand.kaist.ac.kr
Department of Materials Science and EngineeringKAIST
2022.05.20 View 16228
Neuromorphic Memory Device Simulates Neurons and Synapses
Simultaneous emulation of neuronal and synaptic properties promotes the development of brain-like artificial intelligence
Researchers have reported a nano-sized neuromorphic memory device that emulates neurons and synapses simultaneously in a unit cell, another step toward completing the goal of neuromorphic computing designed to rigorously mimic the human brain with semiconductor devices.
Neuromorphic computing aims to realize artificial intelligence (AI) by mimicking the mechanisms of neurons and synapses that make up the human brain. Inspired by the cognitive functions of the human brain that current computers cannot provide, neuromorphic devices have been widely investigated. However, current Complementary Metal-Oxide Semiconductor (CMOS)-based neuromorphic circuits simply connect artificial neurons and synapses without synergistic interactions, and the concomitant implementation of neurons and synapses still remains a challenge. To address these issues, a research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering implemented the biological working mechanisms of humans by introducing the neuron-synapse interactions in a single memory cell, rather than the conventional approach of electrically connecting artificial neuronal and synaptic devices.
Similar to commercial graphics cards, the artificial synaptic devices previously studied often used to accelerate parallel computations, which shows clear differences from the operational mechanisms of the human brain. The research team implemented the synergistic interactions between neurons and synapses in the neuromorphic memory device, emulating the mechanisms of the biological neural network. In addition, the developed neuromorphic device can replace complex CMOS neuron circuits with a single device, providing high scalability and cost efficiency.
The human brain consists of a complex network of 100 billion neurons and 100 trillion synapses. The functions and structures of neurons and synapses can flexibly change according to the external stimuli, adapting to the surrounding environment. The research team developed a neuromorphic device in which short-term and long-term memories coexist using volatile and non-volatile memory devices that mimic the characteristics of neurons and synapses, respectively. A threshold switch device is used as volatile memory and phase-change memory is used as a non-volatile device. Two thin-film devices are integrated without intermediate electrodes, implementing the functional adaptability of neurons and synapses in the neuromorphic memory.
Professor Keon Jae Lee explained, "Neurons and synapses interact with each other to establish cognitive functions such as memory and learning, so simulating both is an essential element for brain-inspired artificial intelligence. The developed neuromorphic memory device also mimics the retraining effect that allows quick learning of the forgotten information by implementing a positive feedback effect between neurons and synapses.”
This result entitled “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse” was published in the May 19, 2022 issue of Nature Communications.
-Publication:Sang Hyun Sung, Tae Jin Kim, Hyera Shin, Tae Hong Im, and Keon Jae Lee (2022) “Simultaneous emulation of synaptic and intrinsic plasticity using a memristive synapse,” Nature Communications May 19, 2022 (DOI: 10.1038/s41467-022-30432-2)
-Profile:Professor Keon Jae Leehttp://fand.kaist.ac.kr
Department of Materials Science and EngineeringKAIST
2022.05.20 View 16228 -
 Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 12331
Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 12331 -
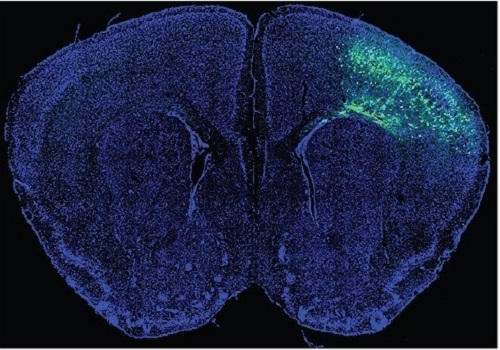 A Mechanism Underlying Most Common Cause of Epileptic Seizures Revealed
An interdisciplinary study shows that neurons carrying somatic mutations in MTOR can lead to focal epileptogenesis via non-cell-autonomous hyperexcitability of nearby nonmutated neurons
During fetal development, cells should migrate to the outer edge of the brain to form critical connections for information transfer and regulation in the body. When even a few cells fail to move to the correct location, the neurons become disorganized and this results in focal cortical dysplasia. This condition is the most common cause of seizures that cannot be controlled with medication in children and the second most common cause in adults.
Now, an interdisciplinary team studying neurogenetics, neural networks, and neurophysiology at KAIST has revealed how dysfunctions in even a small percentage of cells can cause disorder across the entire brain. They published their results on June 28 in Annals of Neurology.
The work builds on a previous finding, also by a KAIST scientists, who found that focal cortical dysplasia was caused by mutations in the cells involved in mTOR, a pathway that regulates signaling between neurons in the brain.
“Only 1 to 2% of neurons carrying mutations in the mTOR signaling pathway that regulates cell signaling in the brain have been found to include seizures in animal models of focal cortical dysplasia,” said Professor Jong-Woo Sohn from the Department of Biological Sciences. “The main challenge of this study was to explain how nearby non-mutated neurons are hyperexcitable.”
Initially, the researchers hypothesized that the mutated cells affected the number of excitatory and inhibitory synapses in all neurons, mutated or not. These neural gates can trigger or halt activity, respectively, in other neurons. Seizures are a result of extreme activity, called hyperexcitability. If the mutated cells upend the balance and result in more excitatory cells, the researchers thought, it made sense that the cells would be more susceptible to hyperexcitability and, as a result, seizures.
“Contrary to our expectations, the synaptic input balance was not changed in either the mutated or non-mutated neurons,” said Professor Jeong Ho Lee from the Graduate School of Medical Science and Engineering. “We turned our attention to a protein overproduced by mutated neurons.”
The protein is adenosine kinase, which lowers the concentration of adenosine. This naturally occurring compound is an anticonvulsant and works to relax vessels. In mice engineered to have focal cortical dysplasia, the researchers injected adenosine to replace the levels lowered by the protein. It worked and the neurons became less excitable.
“We demonstrated that augmentation of adenosine signaling could attenuate the excitability of non-mutated neurons,” said Professor Se-Bum Paik from the Department of Bio and Brain Engineering.
The effect on the non-mutated neurons was the surprising part, according to Paik. “The seizure-triggering hyperexcitability originated not in the mutation-carrying neurons, but instead in the nearby non-mutated neurons,” he said.
The mutated neurons excreted more adenosine kinase, reducing the adenosine levels in the local environment of all the cells. With less adenosine, the non-mutated neurons became hyperexcitable, leading to seizures.
“While we need further investigate into the relationship between the concentration of adenosine and the increased excitation of nearby neurons, our results support the medical use of drugs to activate adenosine signaling as a possible treatment pathway for focal cortical dysplasia,” Professor Lee said.
The Suh Kyungbae Foundation, the Korea Health Technology Research and Development Project, the Ministry of Health & Welfare, and the National Research Foundation in Korea funded this work.
-Publication:Koh, H.Y., Jang, J., Ju, S.H., Kim, R., Cho, G.-B., Kim, D.S., Sohn, J.-W., Paik, S.-B. and Lee, J.H. (2021), ‘Non–Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia’ Annals of Neurology, 90: 285 299. (https://doi.org/10.1002/ana.26149)
-ProfileProfessor Jeong Ho Lee Translational Neurogenetics Labhttps://tnl.kaist.ac.kr/ Graduate School of Medical Science and Engineering KAIST
Professor Se-Bum Paik Visual System and Neural Network Laboratory http://vs.kaist.ac.kr/ Department of Bio and Brain EngineeringKAIST
Professor Jong-Woo Sohn Laboratory for Neurophysiology, https://sites.google.com/site/sohnlab2014/home Department of Biological SciencesKAIST
Dr. Hyun Yong Koh Translational Neurogenetics LabGraduate School of Medical Science and EngineeringKAIST
Dr. Jaeson Jang Ph.D.Visual System and Neural Network LaboratoryDepartment of Bio and Brain Engineering KAIST
Sang Hyeon Ju M.D.Laboratory for NeurophysiologyDepartment of Biological SciencesKAIST
2021.08.26 View 15866
A Mechanism Underlying Most Common Cause of Epileptic Seizures Revealed
An interdisciplinary study shows that neurons carrying somatic mutations in MTOR can lead to focal epileptogenesis via non-cell-autonomous hyperexcitability of nearby nonmutated neurons
During fetal development, cells should migrate to the outer edge of the brain to form critical connections for information transfer and regulation in the body. When even a few cells fail to move to the correct location, the neurons become disorganized and this results in focal cortical dysplasia. This condition is the most common cause of seizures that cannot be controlled with medication in children and the second most common cause in adults.
Now, an interdisciplinary team studying neurogenetics, neural networks, and neurophysiology at KAIST has revealed how dysfunctions in even a small percentage of cells can cause disorder across the entire brain. They published their results on June 28 in Annals of Neurology.
The work builds on a previous finding, also by a KAIST scientists, who found that focal cortical dysplasia was caused by mutations in the cells involved in mTOR, a pathway that regulates signaling between neurons in the brain.
“Only 1 to 2% of neurons carrying mutations in the mTOR signaling pathway that regulates cell signaling in the brain have been found to include seizures in animal models of focal cortical dysplasia,” said Professor Jong-Woo Sohn from the Department of Biological Sciences. “The main challenge of this study was to explain how nearby non-mutated neurons are hyperexcitable.”
Initially, the researchers hypothesized that the mutated cells affected the number of excitatory and inhibitory synapses in all neurons, mutated or not. These neural gates can trigger or halt activity, respectively, in other neurons. Seizures are a result of extreme activity, called hyperexcitability. If the mutated cells upend the balance and result in more excitatory cells, the researchers thought, it made sense that the cells would be more susceptible to hyperexcitability and, as a result, seizures.
“Contrary to our expectations, the synaptic input balance was not changed in either the mutated or non-mutated neurons,” said Professor Jeong Ho Lee from the Graduate School of Medical Science and Engineering. “We turned our attention to a protein overproduced by mutated neurons.”
The protein is adenosine kinase, which lowers the concentration of adenosine. This naturally occurring compound is an anticonvulsant and works to relax vessels. In mice engineered to have focal cortical dysplasia, the researchers injected adenosine to replace the levels lowered by the protein. It worked and the neurons became less excitable.
“We demonstrated that augmentation of adenosine signaling could attenuate the excitability of non-mutated neurons,” said Professor Se-Bum Paik from the Department of Bio and Brain Engineering.
The effect on the non-mutated neurons was the surprising part, according to Paik. “The seizure-triggering hyperexcitability originated not in the mutation-carrying neurons, but instead in the nearby non-mutated neurons,” he said.
The mutated neurons excreted more adenosine kinase, reducing the adenosine levels in the local environment of all the cells. With less adenosine, the non-mutated neurons became hyperexcitable, leading to seizures.
“While we need further investigate into the relationship between the concentration of adenosine and the increased excitation of nearby neurons, our results support the medical use of drugs to activate adenosine signaling as a possible treatment pathway for focal cortical dysplasia,” Professor Lee said.
The Suh Kyungbae Foundation, the Korea Health Technology Research and Development Project, the Ministry of Health & Welfare, and the National Research Foundation in Korea funded this work.
-Publication:Koh, H.Y., Jang, J., Ju, S.H., Kim, R., Cho, G.-B., Kim, D.S., Sohn, J.-W., Paik, S.-B. and Lee, J.H. (2021), ‘Non–Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia’ Annals of Neurology, 90: 285 299. (https://doi.org/10.1002/ana.26149)
-ProfileProfessor Jeong Ho Lee Translational Neurogenetics Labhttps://tnl.kaist.ac.kr/ Graduate School of Medical Science and Engineering KAIST
Professor Se-Bum Paik Visual System and Neural Network Laboratory http://vs.kaist.ac.kr/ Department of Bio and Brain EngineeringKAIST
Professor Jong-Woo Sohn Laboratory for Neurophysiology, https://sites.google.com/site/sohnlab2014/home Department of Biological SciencesKAIST
Dr. Hyun Yong Koh Translational Neurogenetics LabGraduate School of Medical Science and EngineeringKAIST
Dr. Jaeson Jang Ph.D.Visual System and Neural Network LaboratoryDepartment of Bio and Brain Engineering KAIST
Sang Hyeon Ju M.D.Laboratory for NeurophysiologyDepartment of Biological SciencesKAIST
2021.08.26 View 15866 -
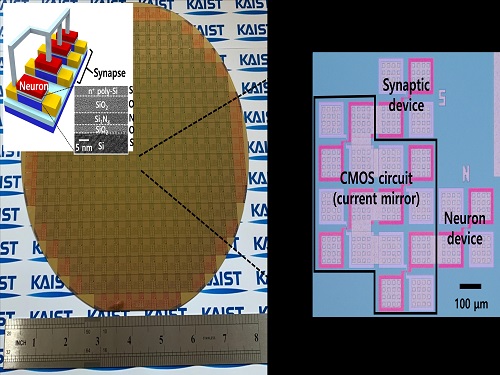 Brain-Inspired Highly Scalable Neuromorphic Hardware Presented
Neurons and synapses based on single transistor can dramatically reduce the hardware cost and accelerate the commercialization of neuromorphic hardware
KAIST researchers fabricated a brain-inspired highly scalable neuromorphic hardware by co-integrating single transistor neurons and synapses. Using standard silicon complementary metal-oxide-semiconductor (CMOS) technology, the neuromorphic hardware is expected to reduce chip cost and simplify fabrication procedures.
The research team led by Yang-Kyu Choi and Sung-Yool Choi produced a neurons and synapses based on single transistor for highly scalable neuromorphic hardware and showed the ability to recognize text and face images. This research was featured in Science Advances on August 4.
Neuromorphic hardware has attracted a great deal of attention because of its artificial intelligence functions, but consuming ultra-low power of less than 20 watts by mimicking the human brain. To make neuromorphic hardware work, a neuron that generates a spike when integrating a certain signal, and a synapse remembering the connection between two neurons are necessary, just like the biological brain. However, since neurons and synapses constructed on digital or analog circuits occupy a large space, there is a limit in terms of hardware efficiency and costs. Since the human brain consists of about 1011 neurons and 1014 synapses, it is necessary to improve the hardware cost in order to apply it to mobile and IoT devices.
To solve the problem, the research team mimicked the behavior of biological neurons and synapses with a single transistor, and co-integrated them onto an 8-inch wafer. The manufactured neuromorphic transistors have the same structure as the transistors for memory and logic that are currently mass-produced. In addition, the neuromorphic transistors proved for the first time that they can be implemented with a ‘Janus structure’ that functions as both neuron and synapse, just like coins have heads and tails.
Professor Yang-Kyu Choi said that this work can dramatically reduce the hardware cost by replacing the neurons and synapses that were based on complex digital and analog circuits with a single transistor. "We have demonstrated that neurons and synapses can be implemented using a single transistor," said Joon-Kyu Han, the first author. "By co-integrating single transistor neurons and synapses on the same wafer using a standard CMOS process, the hardware cost of the neuromorphic hardware has been improved, which will accelerate the commercialization of neuromorphic hardware,” Han added.This research was supported by the National Research Foundation (NRF) and IC Design Education Center (IDEC).
-PublicationJoon-Kyu Han, Sung-Yool Choi, Yang-Kyu Choi, et al.“Cointegration of single-transistor neurons and synapses by nanoscale CMOS fabrication for highly scalable neuromorphic hardware,” Science Advances (DOI: 10.1126/sciadv.abg8836)
-ProfileProfessor Yang-Kyu ChoiNano-Oriented Bio-Electronics Labhttps://sites.google.com/view/nobelab/
School of Electrical EngineeringKAIST
Professor Sung-Yool ChoiMolecular and Nano Device Laboratoryhttps://www.mndl.kaist.ac.kr/
School of Electrical EngineeringKAIST
2021.08.05 View 13245
Brain-Inspired Highly Scalable Neuromorphic Hardware Presented
Neurons and synapses based on single transistor can dramatically reduce the hardware cost and accelerate the commercialization of neuromorphic hardware
KAIST researchers fabricated a brain-inspired highly scalable neuromorphic hardware by co-integrating single transistor neurons and synapses. Using standard silicon complementary metal-oxide-semiconductor (CMOS) technology, the neuromorphic hardware is expected to reduce chip cost and simplify fabrication procedures.
The research team led by Yang-Kyu Choi and Sung-Yool Choi produced a neurons and synapses based on single transistor for highly scalable neuromorphic hardware and showed the ability to recognize text and face images. This research was featured in Science Advances on August 4.
Neuromorphic hardware has attracted a great deal of attention because of its artificial intelligence functions, but consuming ultra-low power of less than 20 watts by mimicking the human brain. To make neuromorphic hardware work, a neuron that generates a spike when integrating a certain signal, and a synapse remembering the connection between two neurons are necessary, just like the biological brain. However, since neurons and synapses constructed on digital or analog circuits occupy a large space, there is a limit in terms of hardware efficiency and costs. Since the human brain consists of about 1011 neurons and 1014 synapses, it is necessary to improve the hardware cost in order to apply it to mobile and IoT devices.
To solve the problem, the research team mimicked the behavior of biological neurons and synapses with a single transistor, and co-integrated them onto an 8-inch wafer. The manufactured neuromorphic transistors have the same structure as the transistors for memory and logic that are currently mass-produced. In addition, the neuromorphic transistors proved for the first time that they can be implemented with a ‘Janus structure’ that functions as both neuron and synapse, just like coins have heads and tails.
Professor Yang-Kyu Choi said that this work can dramatically reduce the hardware cost by replacing the neurons and synapses that were based on complex digital and analog circuits with a single transistor. "We have demonstrated that neurons and synapses can be implemented using a single transistor," said Joon-Kyu Han, the first author. "By co-integrating single transistor neurons and synapses on the same wafer using a standard CMOS process, the hardware cost of the neuromorphic hardware has been improved, which will accelerate the commercialization of neuromorphic hardware,” Han added.This research was supported by the National Research Foundation (NRF) and IC Design Education Center (IDEC).
-PublicationJoon-Kyu Han, Sung-Yool Choi, Yang-Kyu Choi, et al.“Cointegration of single-transistor neurons and synapses by nanoscale CMOS fabrication for highly scalable neuromorphic hardware,” Science Advances (DOI: 10.1126/sciadv.abg8836)
-ProfileProfessor Yang-Kyu ChoiNano-Oriented Bio-Electronics Labhttps://sites.google.com/view/nobelab/
School of Electrical EngineeringKAIST
Professor Sung-Yool ChoiMolecular and Nano Device Laboratoryhttps://www.mndl.kaist.ac.kr/
School of Electrical EngineeringKAIST
2021.08.05 View 13245 -
 A Single, Master Switch for Sugar Levels?
When a fly eats sugar, a single brain cell sends simultaneous messages to stimulate one hormone and inhibit another to control glucose levels in the body. Further research into this control system with remarkable precision could shed light on the neural mechanisms of diabetes and obesity in humans .
A single neuron appears to monitor and control sugar levels in the fly body, according to research published this week in Nature. This new insight into the mechanisms in the fly brain that maintain a balance of two key hormones controlling glucose levels, insulin and glucagon, can provide a framework for understanding diabetes and obesity in humans.
Neurons that sense and respond to glucose were identified more than 50 years ago, but what they do in our body has remained unclear. Researchers at the Korea Advanced Institute of Science and Technology (KAIST) and New York University School of Medicine have now found a single “glucose-sensing neuron” that appears to be the master controller in Drosophila, the vinegar fly, for maintaining an ideal glucose balance, called homeostasis.
Professor Greg Seong-Bae Suh, Dr. Yangkyun Oh and colleagues identified a key neuron that is excited by glucose, which they called CN neuron. This CN neuron has a unique shape – it has an axon (which is used to transmit information to downstream cells) that is bifurcated. One branch projects to insulin-producing cells, and sends a signal triggering the secretion of the insulin equivalent in flies. The other branch projects to glucagon-producing cells and sends a signal inhibiting the secretion of the glucagon equivalent.
When flies consume food, the levels of glucose in their body increase; this excites the CN neuron, which fires the simultaneous signals to stimulate insulin and inhibit glucagon secretion, thereby maintaining the appropriate balance between the hormones and sugar in the blood. The researchers were able to see this happening in the brain in real time by using a combination of cutting-edge fluorescent calcium imaging technology, as well as measuring hormone and sugar levels and applying highly sophisticated molecular genetic techniques.
When flies were not fed, however, the researchers observed a reduction in the activity of CN neuron, a reduction in insulin secretion and an increase in glucagon secretion. These findings indicate that these key hormones are under the direct control of the glucose-sensing neuron. Furthermore, when they silenced the CN neuron rendering dysfunctional CN neuron in flies, these animals experienced an imbalance, resulting in hyperglycemia – high levels of sugars in the blood, similar to what is observed in diabetes in humans. This further suggests that the CN neuron is critical to maintaining glucose homeostasis in animals.
While further research is required to investigate this process in humans, Suh notes this is a significant step forward in the fields of both neurobiology and endocrinology.
“This work lays the foundation for translational research to better understand how this delicate regulatory process is affected by diabetes, obesity, excessive nutrition and diets high in sugar,” Suh said.
Profile: Greg Seong-Bae Suh
seongbaesuh@kaist.ac.kr
Professor Department of Biological Sciences
KAIST
(Figure: A single glucose-excited CN neuron extends bifurcated axonal branches,
one of which innervates insulin producing cells and stimulates their activity an the other axonal branch projects to glucagon producing cells and inhibits their activity.)
2019.10.24 View 19249
A Single, Master Switch for Sugar Levels?
When a fly eats sugar, a single brain cell sends simultaneous messages to stimulate one hormone and inhibit another to control glucose levels in the body. Further research into this control system with remarkable precision could shed light on the neural mechanisms of diabetes and obesity in humans .
A single neuron appears to monitor and control sugar levels in the fly body, according to research published this week in Nature. This new insight into the mechanisms in the fly brain that maintain a balance of two key hormones controlling glucose levels, insulin and glucagon, can provide a framework for understanding diabetes and obesity in humans.
Neurons that sense and respond to glucose were identified more than 50 years ago, but what they do in our body has remained unclear. Researchers at the Korea Advanced Institute of Science and Technology (KAIST) and New York University School of Medicine have now found a single “glucose-sensing neuron” that appears to be the master controller in Drosophila, the vinegar fly, for maintaining an ideal glucose balance, called homeostasis.
Professor Greg Seong-Bae Suh, Dr. Yangkyun Oh and colleagues identified a key neuron that is excited by glucose, which they called CN neuron. This CN neuron has a unique shape – it has an axon (which is used to transmit information to downstream cells) that is bifurcated. One branch projects to insulin-producing cells, and sends a signal triggering the secretion of the insulin equivalent in flies. The other branch projects to glucagon-producing cells and sends a signal inhibiting the secretion of the glucagon equivalent.
When flies consume food, the levels of glucose in their body increase; this excites the CN neuron, which fires the simultaneous signals to stimulate insulin and inhibit glucagon secretion, thereby maintaining the appropriate balance between the hormones and sugar in the blood. The researchers were able to see this happening in the brain in real time by using a combination of cutting-edge fluorescent calcium imaging technology, as well as measuring hormone and sugar levels and applying highly sophisticated molecular genetic techniques.
When flies were not fed, however, the researchers observed a reduction in the activity of CN neuron, a reduction in insulin secretion and an increase in glucagon secretion. These findings indicate that these key hormones are under the direct control of the glucose-sensing neuron. Furthermore, when they silenced the CN neuron rendering dysfunctional CN neuron in flies, these animals experienced an imbalance, resulting in hyperglycemia – high levels of sugars in the blood, similar to what is observed in diabetes in humans. This further suggests that the CN neuron is critical to maintaining glucose homeostasis in animals.
While further research is required to investigate this process in humans, Suh notes this is a significant step forward in the fields of both neurobiology and endocrinology.
“This work lays the foundation for translational research to better understand how this delicate regulatory process is affected by diabetes, obesity, excessive nutrition and diets high in sugar,” Suh said.
Profile: Greg Seong-Bae Suh
seongbaesuh@kaist.ac.kr
Professor Department of Biological Sciences
KAIST
(Figure: A single glucose-excited CN neuron extends bifurcated axonal branches,
one of which innervates insulin producing cells and stimulates their activity an the other axonal branch projects to glucagon producing cells and inhibits their activity.)
2019.10.24 View 19249