Bio
-
 KAIST Successfully Implements 3D Brain-Mimicking Platform with 6x Higher Precision
<(From left) Dr. Dongjo Yoon, Professor Je-Kyun Park from the Department of Bio and Brain Engineering, (upper right) Professor Yoonkey Nam, Dr. Soo Jee Kim>
Existing three-dimensional (3D) neuronal culture technology has limitations in brain research due to the difficulty of precisely replicating the brain's complex multilayered structure and the lack of a platform that can simultaneously analyze both structure and function. A KAIST research team has successfully developed an integrated platform that can implement brain-like layered neuronal structures using 3D printing technology and precisely measure neuronal activity within them.
KAIST (President Kwang Hyung Lee) announced on the 16th of July that a joint research team led by Professors Je-Kyun Park and Yoonkey Nam from the Department of Bio and Brain Engineering has developed an integrated platform capable of fabricating high-resolution 3D multilayer neuronal networks using low-viscosity natural hydrogels with mechanical properties similar to brain tissue, and simultaneously analyzing their structural and functional connectivity.
Conventional bioprinting technology uses high-viscosity bioinks for structural stability, but this limits neuronal proliferation and neurite growth. Conversely, neural cell-friendly low-viscosity hydrogels are difficult to precisely pattern, leading to a fundamental trade-off between structural stability and biological function. The research team completed a sophisticated and stable brain-mimicking platform by combining three key technologies that enable the precise creation of brain structure with dilute gels, accurate alignment between layers, and simultaneous observation of neuronal activity.
The three core technologies are: ▲ 'Capillary Pinning Effect' technology, which enables the dilute gel (hydrogel) to adhere firmly to a stainless steel mesh (micromesh) to prevent it from flowing, thereby reproducing brain structures with six times greater precision (resolution of 500 μm or less) than conventional methods; ▲ the '3D Printing Aligner,' a cylindrical design that ensures the printed layers are precisely stacked without misalignment, guaranteeing the accurate assembly of multilayer structures and stable integration with microelectrode chips; and ▲ 'Dual-mode Analysis System' technology, which simultaneously measures electrical signals from below and observes cell activity with light (calcium imaging) from above, allowing for the simultaneous verification of the functional operation of interlayer connections through multiple methods.
< Figure 1. Platform integrating brain-structure-mimicking neural network model construction and functional measurement technology>
The research team successfully implemented a three-layered mini-brain structure using 3D printing with a fibrin hydrogel, which has elastic properties similar to those of the brain, and experimentally verified the process of actual neural cells transmitting and receiving signals within it.
Cortical neurons were placed in the upper and lower layers, while the middle layer was left empty but designed to allow neurons to penetrate and connect through it. Electrical signals were measured from the lower layer using a microsensor (electrode chip), and cell activity was observed from the upper layer using light (calcium imaging). The results showed that when electrical stimulation was applied, neural cells in both upper and lower layers responded simultaneously. When a synapse-blocking agent (synaptic blocker) was introduced, the response decreased, proving that the neural cells were genuinely connected and transmitting signals.
Professor Je-Kyun Park of KAIST explained, "This research is a joint development achievement of an integrated platform that can simultaneously reproduce the complex multilayered structure and function of brain tissue. Compared to existing technologies where signal measurement was impossible for more than 14 days, this platform maintains a stable microelectrode chip interface for over 27 days, allowing the real-time analysis of structure-function relationships. It can be utilized in various brain research fields such as neurological disease modeling, brain function research, neurotoxicity assessment, and neuroprotective drug screening in the future."
<Figure 2. Integration process of stacked bioprinting technology and microelectrode chip>
The research, in which Dr. Soo Jee Kim and Dr. Dongjo Yoon from KAIST's Department of Bio and Brain Engineering participated as co-first authors, was published online in the international journal 'Biosensors and Bioelectronics' on June 11, 2025.
※Paper: Hybrid biofabrication of multilayered 3D neuronal networks with structural and functional interlayer connectivity
※DOI: https://doi.org/10.1016/j.bios.2025.117688
2025.07.16 View 81
KAIST Successfully Implements 3D Brain-Mimicking Platform with 6x Higher Precision
<(From left) Dr. Dongjo Yoon, Professor Je-Kyun Park from the Department of Bio and Brain Engineering, (upper right) Professor Yoonkey Nam, Dr. Soo Jee Kim>
Existing three-dimensional (3D) neuronal culture technology has limitations in brain research due to the difficulty of precisely replicating the brain's complex multilayered structure and the lack of a platform that can simultaneously analyze both structure and function. A KAIST research team has successfully developed an integrated platform that can implement brain-like layered neuronal structures using 3D printing technology and precisely measure neuronal activity within them.
KAIST (President Kwang Hyung Lee) announced on the 16th of July that a joint research team led by Professors Je-Kyun Park and Yoonkey Nam from the Department of Bio and Brain Engineering has developed an integrated platform capable of fabricating high-resolution 3D multilayer neuronal networks using low-viscosity natural hydrogels with mechanical properties similar to brain tissue, and simultaneously analyzing their structural and functional connectivity.
Conventional bioprinting technology uses high-viscosity bioinks for structural stability, but this limits neuronal proliferation and neurite growth. Conversely, neural cell-friendly low-viscosity hydrogels are difficult to precisely pattern, leading to a fundamental trade-off between structural stability and biological function. The research team completed a sophisticated and stable brain-mimicking platform by combining three key technologies that enable the precise creation of brain structure with dilute gels, accurate alignment between layers, and simultaneous observation of neuronal activity.
The three core technologies are: ▲ 'Capillary Pinning Effect' technology, which enables the dilute gel (hydrogel) to adhere firmly to a stainless steel mesh (micromesh) to prevent it from flowing, thereby reproducing brain structures with six times greater precision (resolution of 500 μm or less) than conventional methods; ▲ the '3D Printing Aligner,' a cylindrical design that ensures the printed layers are precisely stacked without misalignment, guaranteeing the accurate assembly of multilayer structures and stable integration with microelectrode chips; and ▲ 'Dual-mode Analysis System' technology, which simultaneously measures electrical signals from below and observes cell activity with light (calcium imaging) from above, allowing for the simultaneous verification of the functional operation of interlayer connections through multiple methods.
< Figure 1. Platform integrating brain-structure-mimicking neural network model construction and functional measurement technology>
The research team successfully implemented a three-layered mini-brain structure using 3D printing with a fibrin hydrogel, which has elastic properties similar to those of the brain, and experimentally verified the process of actual neural cells transmitting and receiving signals within it.
Cortical neurons were placed in the upper and lower layers, while the middle layer was left empty but designed to allow neurons to penetrate and connect through it. Electrical signals were measured from the lower layer using a microsensor (electrode chip), and cell activity was observed from the upper layer using light (calcium imaging). The results showed that when electrical stimulation was applied, neural cells in both upper and lower layers responded simultaneously. When a synapse-blocking agent (synaptic blocker) was introduced, the response decreased, proving that the neural cells were genuinely connected and transmitting signals.
Professor Je-Kyun Park of KAIST explained, "This research is a joint development achievement of an integrated platform that can simultaneously reproduce the complex multilayered structure and function of brain tissue. Compared to existing technologies where signal measurement was impossible for more than 14 days, this platform maintains a stable microelectrode chip interface for over 27 days, allowing the real-time analysis of structure-function relationships. It can be utilized in various brain research fields such as neurological disease modeling, brain function research, neurotoxicity assessment, and neuroprotective drug screening in the future."
<Figure 2. Integration process of stacked bioprinting technology and microelectrode chip>
The research, in which Dr. Soo Jee Kim and Dr. Dongjo Yoon from KAIST's Department of Bio and Brain Engineering participated as co-first authors, was published online in the international journal 'Biosensors and Bioelectronics' on June 11, 2025.
※Paper: Hybrid biofabrication of multilayered 3D neuronal networks with structural and functional interlayer connectivity
※DOI: https://doi.org/10.1016/j.bios.2025.117688
2025.07.16 View 81 -
 A KAIST Team Engineers a Microbial Platform for Efficient Lutein Production
<(From Left) Ph.D. Candidate Hyunmin Eun, Distinguished Professor Sang Yup Lee, , Dr. Cindy Pricilia Surya Prabowo>
The application of systems metabolic engineering strategies, along with the construction of an electron channeling system, has enabled the first gram-per-liter scale production of lutein from Corynebacterium glutamicum, providing a viable alternative to plant-derived lutein production.
A research group at KAIST has successfully engineered a microbial strain capable of producing lutein at industrially relevant levels. The team, led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering, developed a novel C. glutamicum strain using systems metabolic engineering strategies to overcome the limitations of previous microbial lutein production efforts. This research is expected to be beneficial for the efficient production of other industrially important natural products used in food, pharmaceuticals, and cosmetics.
Lutein is a xanthophyll carotenoid found in egg yolk, fruits, and vegetables, known for its role in protecting our eyes from oxidative stress and reducing the risk of macular degeneration and cataracts. Currently, commercial lutein is predominantly extracted from marigold flowers; however, this approach has several drawbacks, including long cultivation times, high labor costs, and inefficient extraction yields, making it economically unfeasible for large-scale production. These challenges have driven the demand for alternative production methods.
To address these issues, KAIST researchers, including Ph.D. Candidate Hyunmin Eun, Dr. Cindy Pricilia Surya Prabowo, and Distinguished Professor Sang Yup Lee, applied systems metabolic engineering strategies to engineer C. glutamicum, a GRAS (Generally Recognized As Safe) microorganism widely used in industrial fermentation. Unlike Escherichia coli, which was previously explored for microbial lutein production, C. glutamicum lacks endotoxins, making it a safer and more viable option for food and pharmaceutical applications.
The team’s work, entitled “Gram-per-litre scale production of lutein by engineered Corynebacterium,” was published in Nature Synthesis on 04 July , 2025.
This research details the high-level production of lutein using glucose as a renewable carbon source via systems metabolic engineering. The team focused on eliminating metabolic bottlenecks that previously limited microbial lutein synthesis. By employing enzyme scaffold-based electron channeling strategies, the researchers improved metabolic flux towards lutein biosynthesis while minimizing unwanted byproducts.
<Lutein production metabolic pathway engineering>
To enhance productivity, bottleneck enzymes within the metabolic pathway were identified and optimized. It was determined that electron-requiring cytochrome P450 enzymes played a major role in limiting lutein biosynthesis. To overcome this limitation, an electron channeling strategy was implemented, where engineered cytochrome P450 enzymes and their reductase partners were spatially organized on synthetic scaffolds, allowing more efficient electron transfer and significantly increasing lutein production.
The engineered C. glutamicum strain was further optimized in fed-batch fermentation, achieving a record-breaking 1.78 g/L of lutein production within 54 hours, with a content of 19.51 mg/gDCW and a productivity of 32.88 mg/L/h—the highest lutein production performance in any host reported to date. This milestone demonstrates the feasibility of replacing plant-based lutein extraction with microbial fermentation technology.
“We can anticipate that this microbial cell factory-based mass production of lutein will be able to replace the current plant extraction-based process,” said Ph.D. Candidate Hyunmin Eun. He emphasized that the integrated metabolic engineering strategies developed in this study could be broadly applied for the efficient production of other valuable natural products used in pharmaceuticals and nutraceuticals.
<Schematic diagram of microbial-based lutein production platform>
“As maintaining good health in an aging society becomes increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other medically and nutritionally significant natural products,” added Distinguished Professor Sang Yup Lee.
This work is supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry project 2022M3J5A1056072 and the Development of Platform Technologies of Microbial Cell Factories for the Next-Generation Biorefineries project 2022M3J5A1056117 from the National Research Foundation supported by the Korean Ministry of Science and ICT.
Source:
Hyunmin Eun (1st), Cindy Pricilia Surya Prabowo (co-1st), and Sang Yup Lee (Corresponding). “Gram-per-litre scale production of lutein by engineered Corynebacterium”. Nature Synthesis (Online published)
For further information:
Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering, KAIST (leesy@kaist.ac.kr, Tel: +82-42-350-3930)
2025.07.14 View 230
A KAIST Team Engineers a Microbial Platform for Efficient Lutein Production
<(From Left) Ph.D. Candidate Hyunmin Eun, Distinguished Professor Sang Yup Lee, , Dr. Cindy Pricilia Surya Prabowo>
The application of systems metabolic engineering strategies, along with the construction of an electron channeling system, has enabled the first gram-per-liter scale production of lutein from Corynebacterium glutamicum, providing a viable alternative to plant-derived lutein production.
A research group at KAIST has successfully engineered a microbial strain capable of producing lutein at industrially relevant levels. The team, led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering, developed a novel C. glutamicum strain using systems metabolic engineering strategies to overcome the limitations of previous microbial lutein production efforts. This research is expected to be beneficial for the efficient production of other industrially important natural products used in food, pharmaceuticals, and cosmetics.
Lutein is a xanthophyll carotenoid found in egg yolk, fruits, and vegetables, known for its role in protecting our eyes from oxidative stress and reducing the risk of macular degeneration and cataracts. Currently, commercial lutein is predominantly extracted from marigold flowers; however, this approach has several drawbacks, including long cultivation times, high labor costs, and inefficient extraction yields, making it economically unfeasible for large-scale production. These challenges have driven the demand for alternative production methods.
To address these issues, KAIST researchers, including Ph.D. Candidate Hyunmin Eun, Dr. Cindy Pricilia Surya Prabowo, and Distinguished Professor Sang Yup Lee, applied systems metabolic engineering strategies to engineer C. glutamicum, a GRAS (Generally Recognized As Safe) microorganism widely used in industrial fermentation. Unlike Escherichia coli, which was previously explored for microbial lutein production, C. glutamicum lacks endotoxins, making it a safer and more viable option for food and pharmaceutical applications.
The team’s work, entitled “Gram-per-litre scale production of lutein by engineered Corynebacterium,” was published in Nature Synthesis on 04 July , 2025.
This research details the high-level production of lutein using glucose as a renewable carbon source via systems metabolic engineering. The team focused on eliminating metabolic bottlenecks that previously limited microbial lutein synthesis. By employing enzyme scaffold-based electron channeling strategies, the researchers improved metabolic flux towards lutein biosynthesis while minimizing unwanted byproducts.
<Lutein production metabolic pathway engineering>
To enhance productivity, bottleneck enzymes within the metabolic pathway were identified and optimized. It was determined that electron-requiring cytochrome P450 enzymes played a major role in limiting lutein biosynthesis. To overcome this limitation, an electron channeling strategy was implemented, where engineered cytochrome P450 enzymes and their reductase partners were spatially organized on synthetic scaffolds, allowing more efficient electron transfer and significantly increasing lutein production.
The engineered C. glutamicum strain was further optimized in fed-batch fermentation, achieving a record-breaking 1.78 g/L of lutein production within 54 hours, with a content of 19.51 mg/gDCW and a productivity of 32.88 mg/L/h—the highest lutein production performance in any host reported to date. This milestone demonstrates the feasibility of replacing plant-based lutein extraction with microbial fermentation technology.
“We can anticipate that this microbial cell factory-based mass production of lutein will be able to replace the current plant extraction-based process,” said Ph.D. Candidate Hyunmin Eun. He emphasized that the integrated metabolic engineering strategies developed in this study could be broadly applied for the efficient production of other valuable natural products used in pharmaceuticals and nutraceuticals.
<Schematic diagram of microbial-based lutein production platform>
“As maintaining good health in an aging society becomes increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other medically and nutritionally significant natural products,” added Distinguished Professor Sang Yup Lee.
This work is supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry project 2022M3J5A1056072 and the Development of Platform Technologies of Microbial Cell Factories for the Next-Generation Biorefineries project 2022M3J5A1056117 from the National Research Foundation supported by the Korean Ministry of Science and ICT.
Source:
Hyunmin Eun (1st), Cindy Pricilia Surya Prabowo (co-1st), and Sang Yup Lee (Corresponding). “Gram-per-litre scale production of lutein by engineered Corynebacterium”. Nature Synthesis (Online published)
For further information:
Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering, KAIST (leesy@kaist.ac.kr, Tel: +82-42-350-3930)
2025.07.14 View 230 -
 KAIST Shows That the Brain Can Distinguish Glucose: Clues to Treat Obesity and Diabetes
<(From left)Prof. Greg S.B Suh, Dr. Jieun Kim, Dr. Shinhye Kim, Researcher Wongyo Jeong)
“How does our brain distinguish glucose from the many nutrients absorbed in the gut?” Starting with this question, a KAIST research team has demonstrated that the brain can selectively recognize specific nutrients—particularly glucose—beyond simply detecting total calorie content. This study is expected to offer a new paradigm for appetite control and the treatment of metabolic diseases.
On the 9th, KAIST (President Kwang Hyung Lee) announced that Professor Greg S.B. Suh’s team in the Department of Biological Sciences, in collaboration with Professor Young-Gyun Park’s team (BarNeuro), Professor Seung-Hee Lee’s team (Department of Biological Sciences), and the Albert Einstein College of Medicine in New York, had identified the existence of a gut-brain circuit that allows animals in a hungry state to selectively detect and prefer glucose in the gut.
Organisms derive energy from various nutrients including sugars, proteins, and fats. Previous studies have shown that total caloric information in the gut suppresses hunger neurons in the hypothalamus to regulate appetite. However, the existence of a gut-brain circuit that specifically responds to glucose and corresponding brain cells had not been demonstrated until now.
In this study, the team successfully identified a “gut-brain circuit” that senses glucose—essential for brain function—and regulates food intake behavior for required nutrients.
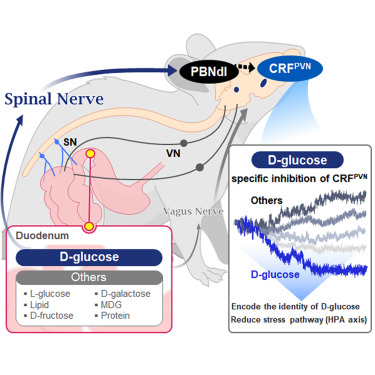
They further proved, for the first time, that this circuit responds within seconds to not only hunger or external stimuli but also to specific caloric nutrients directly introduced into the small intestine, particularly D-glucose, through the activity of “CRF neurons*” in the brain’s hypothalamus.
*CRF neurons: These neurons secrete corticotropin-releasing factor (CRF) in the hypothalamus and are central to the hypothalamic-pituitary-adrenal (HPA) axis, the body’s core physiological system for responding to stress. CRF neurons are known to regulate neuroendocrine balance in response to stress stimuli.
Using optogenetics to precisely track neural activity in real time, the researchers injected various nutrients—D-glucose, L-glucose, amino acids, and fats—directly into the small intestines of mice and observed the results.
They discovered that among the CRF neurons located in the paraventricular nucleus (PVN)* of the hypothalamus, only those specific to D-glucose showed selective responses. These neurons did not respond—or showed inverse reactions—to other sugars or to proteins and fats. This is the first demonstration that single neurons in the brain can guide nutrient-specific responses depending on gut nutrient influx.
*PVN (Paraventricular Nucleus): A key nucleus within the hypothalamus responsible for maintaining bodily homeostasis.
The team also revealed that glucose-sensing signals in the small intestine are transmitted via the spinal cord to the dorsolateral parabrachial nucleus (PBNdl) of the brain, and from there to CRF neurons in the PVN. In contrast, signals for amino acids and fats are transmitted to the brain through the vagus nerve, a different pathway.
In optogenetic inhibition experiments, suppressing CRF neurons in fasting mice eliminated their preference for glucose, proving that this circuit is essential for glucose-specific nutrient preference.
This study was inspired by Professor Suh’s earlier research at NYU using fruit flies, where he identified “DH44 neurons” that selectively detect glucose and sugar in the gut. Based on the hypothesis that hypothalamic neurons in mammals would show similar functional responses to glucose, the current study was launched.
To test this hypothesis, Dr. Jineun Kim (KAIST Ph.D. graduate, now at Caltech) demonstrated during her doctoral research that hungry mice preferred glucose among various intragastrically infused nutrients and that CRF neurons exhibited rapid and specific responses.
Along with Wongyo Jung (KAIST B.S. graduate, now Ph.D. student at Caltech), they modeled and experimentally confirmed the critical role of CRF neurons. Dr. Shinhye Kim, through collaboration, revealed that specific spinal neurons play a key role in conveying intestinal nutrient information to the brain.
Dr. Jineun Kim and Dr. Shinhye Kim said, “This study started from a simple but fundamental question—‘How does the brain distinguish glucose from various nutrients absorbed in the gut?’ We have shown that spinal-based gut-brain circuits play a central role in energy metabolism and homeostasis by transmitting specific gut nutrient signals to the brain.”
Professor Suh added, “By identifying a gut-brain pathway specialized for glucose, this research offers a new therapeutic target for metabolic diseases such as obesity and diabetes. Our future research will explore similar circuits for sensing other essential nutrients like amino acids and fats and their interaction mechanisms.”
Ph.D. student Jineun Kim, Dr. Shinhye Kim, and student Wongyo Jung (co-first authors) contributed to this study, which was published online in the international journal Neuron on June 20, 2025.
※ Paper Title: Encoding the glucose identity by discrete hypothalamic neurons via the gut-brain axis ※ DOI: https://doi.org/10.1016/j.neuron.2025.05.024
This study was supported by the Samsung Science & Technology Foundation, the National Research Foundation of Korea (NRF) Leader Research Program, the POSCO Cheongam Science Fellowship, the Asan Foundation Biomedical Science Scholarship, the Institute for Basic Science (IBS), and the KAIST KAIX program.
2025.07.09 View 267
KAIST Shows That the Brain Can Distinguish Glucose: Clues to Treat Obesity and Diabetes
<(From left)Prof. Greg S.B Suh, Dr. Jieun Kim, Dr. Shinhye Kim, Researcher Wongyo Jeong)
“How does our brain distinguish glucose from the many nutrients absorbed in the gut?” Starting with this question, a KAIST research team has demonstrated that the brain can selectively recognize specific nutrients—particularly glucose—beyond simply detecting total calorie content. This study is expected to offer a new paradigm for appetite control and the treatment of metabolic diseases.
On the 9th, KAIST (President Kwang Hyung Lee) announced that Professor Greg S.B. Suh’s team in the Department of Biological Sciences, in collaboration with Professor Young-Gyun Park’s team (BarNeuro), Professor Seung-Hee Lee’s team (Department of Biological Sciences), and the Albert Einstein College of Medicine in New York, had identified the existence of a gut-brain circuit that allows animals in a hungry state to selectively detect and prefer glucose in the gut.
Organisms derive energy from various nutrients including sugars, proteins, and fats. Previous studies have shown that total caloric information in the gut suppresses hunger neurons in the hypothalamus to regulate appetite. However, the existence of a gut-brain circuit that specifically responds to glucose and corresponding brain cells had not been demonstrated until now.
In this study, the team successfully identified a “gut-brain circuit” that senses glucose—essential for brain function—and regulates food intake behavior for required nutrients.
They further proved, for the first time, that this circuit responds within seconds to not only hunger or external stimuli but also to specific caloric nutrients directly introduced into the small intestine, particularly D-glucose, through the activity of “CRF neurons*” in the brain’s hypothalamus.
*CRF neurons: These neurons secrete corticotropin-releasing factor (CRF) in the hypothalamus and are central to the hypothalamic-pituitary-adrenal (HPA) axis, the body’s core physiological system for responding to stress. CRF neurons are known to regulate neuroendocrine balance in response to stress stimuli.
Using optogenetics to precisely track neural activity in real time, the researchers injected various nutrients—D-glucose, L-glucose, amino acids, and fats—directly into the small intestines of mice and observed the results.
They discovered that among the CRF neurons located in the paraventricular nucleus (PVN)* of the hypothalamus, only those specific to D-glucose showed selective responses. These neurons did not respond—or showed inverse reactions—to other sugars or to proteins and fats. This is the first demonstration that single neurons in the brain can guide nutrient-specific responses depending on gut nutrient influx.
*PVN (Paraventricular Nucleus): A key nucleus within the hypothalamus responsible for maintaining bodily homeostasis.
The team also revealed that glucose-sensing signals in the small intestine are transmitted via the spinal cord to the dorsolateral parabrachial nucleus (PBNdl) of the brain, and from there to CRF neurons in the PVN. In contrast, signals for amino acids and fats are transmitted to the brain through the vagus nerve, a different pathway.
In optogenetic inhibition experiments, suppressing CRF neurons in fasting mice eliminated their preference for glucose, proving that this circuit is essential for glucose-specific nutrient preference.
This study was inspired by Professor Suh’s earlier research at NYU using fruit flies, where he identified “DH44 neurons” that selectively detect glucose and sugar in the gut. Based on the hypothesis that hypothalamic neurons in mammals would show similar functional responses to glucose, the current study was launched.
To test this hypothesis, Dr. Jineun Kim (KAIST Ph.D. graduate, now at Caltech) demonstrated during her doctoral research that hungry mice preferred glucose among various intragastrically infused nutrients and that CRF neurons exhibited rapid and specific responses.
Along with Wongyo Jung (KAIST B.S. graduate, now Ph.D. student at Caltech), they modeled and experimentally confirmed the critical role of CRF neurons. Dr. Shinhye Kim, through collaboration, revealed that specific spinal neurons play a key role in conveying intestinal nutrient information to the brain.
Dr. Jineun Kim and Dr. Shinhye Kim said, “This study started from a simple but fundamental question—‘How does the brain distinguish glucose from various nutrients absorbed in the gut?’ We have shown that spinal-based gut-brain circuits play a central role in energy metabolism and homeostasis by transmitting specific gut nutrient signals to the brain.”
Professor Suh added, “By identifying a gut-brain pathway specialized for glucose, this research offers a new therapeutic target for metabolic diseases such as obesity and diabetes. Our future research will explore similar circuits for sensing other essential nutrients like amino acids and fats and their interaction mechanisms.”
Ph.D. student Jineun Kim, Dr. Shinhye Kim, and student Wongyo Jung (co-first authors) contributed to this study, which was published online in the international journal Neuron on June 20, 2025.
※ Paper Title: Encoding the glucose identity by discrete hypothalamic neurons via the gut-brain axis ※ DOI: https://doi.org/10.1016/j.neuron.2025.05.024
This study was supported by the Samsung Science & Technology Foundation, the National Research Foundation of Korea (NRF) Leader Research Program, the POSCO Cheongam Science Fellowship, the Asan Foundation Biomedical Science Scholarship, the Institute for Basic Science (IBS), and the KAIST KAIX program.
2025.07.09 View 267 -
 KAIST Develops Novel Candidiasis Treatment Overcoming Side Effects and Resistance
<(From left) Ph. D Candidate Ju Yeon Chung, Prof.Hyun Jung Chung, Ph.D candidate Seungju Yang, Ph.D candidate Ayoung Park, Dr. Yoon-Kyoung Hong from Asan Medical Center, Prof. Yong Pil Chong, Dr. Eunhee Jeon>
Candida, a type of fungus, which can spread throughout the body via the bloodstream, leading to organ damage and sepsis. Recently, the incidence of candidiasis has surged due to the increase in immunosuppressive therapies, medical implants, and transplantation. Korean researchers have successfully developed a next-generation treatment that, unlike existing antifungals, selectively acts only on Candida, achieving both high therapeutic efficacy and low side effects simultaneously.
KAIST (President Kwang Hyung Lee) announced on the 8th that a research team led by Professor Hyun-Jung Chung of the Department of Biological Sciences, in collaboration with Professor Yong Pil Jeong's team at Asan Medical Center, developed a gene-based nanotherapy (FTNx) that simultaneously inhibits two key enzymes in the Candida cell wall.
Current antifungal drugs for Candida have low target selectivity, which can affect human cells. Furthermore, their therapeutic efficacy is gradually decreasing due to the emergence of new resistant strains. Especially for immunocompromised patients, the infection progresses rapidly and has a poor prognosis, making the development of new treatments to overcome the limitations of existing therapies urgent.
The developed treatment can be administered systemically, and by combining gene suppression technology with nanomaterial technology, it effectively overcomes the structural limitations of existing compound-based drugs and successfully achieves selective treatment against only Candida.
The research team created a gold nanoparticle-based complex loaded with short DNA fragments called antisense oligonucleotides (ASO), which simultaneously target two crucial enzymes—β-1,3-glucan synthase (FKS1) and chitin synthase (CHS3)—important for forming the cell wall of the Candida fungus.
By applying a surface coating technology that binds to a specific glycolipid structure (a structure combining sugar and fat) on the Candida cell wall, a targeted delivery device was implemented. This successfully achieved a precise targeting effect, ensuring the complex is not delivered to human cells at all but acts selectively only on Candida.
<Figure 1: Overview of antifungal therapy design and experimental approach>
This complex, after entering Candida cells, cleaves the mRNA produced by the FKS1 and CHS3 genes, thereby inhibiting translation and simultaneously blocking the synthesis of cell wall components β-1,3-glucan and chitin. As a result, the
Candida cell wall loses its structural stability and collapses, suppressing bacterial survival and proliferation.
In fact, experiments using a systemic candidiasis model in mice confirmed the therapeutic effect: a significant reduction in
Candida count in the organs, normalization of immune responses, and a notable increase in survival rates were observed in the treated group.
Professor Hyun-Jung Chung, who led the research, stated, "This study presents a method to overcome the issues of human toxicity and drug resistance spread with existing treatments, marking an important turning point by demonstrating the applicability of gene therapy for systemic infections". She added, "We plan to continue research on optimizing administration methods and verifying toxicity for future clinical application."
This research involved Ju Yeon Chung and Yoon-Kyoung Hong as co-first authors , and was published in the international journal 'Nature Communications' on July 1st.
Paper Title: Effective treatment of systemic candidiasis by synergistic targeting of cell wall synthesis
DOI: 10.1038/s41467-025-60684-7
This research was supported by the Ministry of Health and Welfare and the National Research Foundation of Korea.
2025.07.08 View 270
KAIST Develops Novel Candidiasis Treatment Overcoming Side Effects and Resistance
<(From left) Ph. D Candidate Ju Yeon Chung, Prof.Hyun Jung Chung, Ph.D candidate Seungju Yang, Ph.D candidate Ayoung Park, Dr. Yoon-Kyoung Hong from Asan Medical Center, Prof. Yong Pil Chong, Dr. Eunhee Jeon>
Candida, a type of fungus, which can spread throughout the body via the bloodstream, leading to organ damage and sepsis. Recently, the incidence of candidiasis has surged due to the increase in immunosuppressive therapies, medical implants, and transplantation. Korean researchers have successfully developed a next-generation treatment that, unlike existing antifungals, selectively acts only on Candida, achieving both high therapeutic efficacy and low side effects simultaneously.
KAIST (President Kwang Hyung Lee) announced on the 8th that a research team led by Professor Hyun-Jung Chung of the Department of Biological Sciences, in collaboration with Professor Yong Pil Jeong's team at Asan Medical Center, developed a gene-based nanotherapy (FTNx) that simultaneously inhibits two key enzymes in the Candida cell wall.
Current antifungal drugs for Candida have low target selectivity, which can affect human cells. Furthermore, their therapeutic efficacy is gradually decreasing due to the emergence of new resistant strains. Especially for immunocompromised patients, the infection progresses rapidly and has a poor prognosis, making the development of new treatments to overcome the limitations of existing therapies urgent.
The developed treatment can be administered systemically, and by combining gene suppression technology with nanomaterial technology, it effectively overcomes the structural limitations of existing compound-based drugs and successfully achieves selective treatment against only Candida.
The research team created a gold nanoparticle-based complex loaded with short DNA fragments called antisense oligonucleotides (ASO), which simultaneously target two crucial enzymes—β-1,3-glucan synthase (FKS1) and chitin synthase (CHS3)—important for forming the cell wall of the Candida fungus.
By applying a surface coating technology that binds to a specific glycolipid structure (a structure combining sugar and fat) on the Candida cell wall, a targeted delivery device was implemented. This successfully achieved a precise targeting effect, ensuring the complex is not delivered to human cells at all but acts selectively only on Candida.
<Figure 1: Overview of antifungal therapy design and experimental approach>
This complex, after entering Candida cells, cleaves the mRNA produced by the FKS1 and CHS3 genes, thereby inhibiting translation and simultaneously blocking the synthesis of cell wall components β-1,3-glucan and chitin. As a result, the
Candida cell wall loses its structural stability and collapses, suppressing bacterial survival and proliferation.
In fact, experiments using a systemic candidiasis model in mice confirmed the therapeutic effect: a significant reduction in
Candida count in the organs, normalization of immune responses, and a notable increase in survival rates were observed in the treated group.
Professor Hyun-Jung Chung, who led the research, stated, "This study presents a method to overcome the issues of human toxicity and drug resistance spread with existing treatments, marking an important turning point by demonstrating the applicability of gene therapy for systemic infections". She added, "We plan to continue research on optimizing administration methods and verifying toxicity for future clinical application."
This research involved Ju Yeon Chung and Yoon-Kyoung Hong as co-first authors , and was published in the international journal 'Nature Communications' on July 1st.
Paper Title: Effective treatment of systemic candidiasis by synergistic targeting of cell wall synthesis
DOI: 10.1038/s41467-025-60684-7
This research was supported by the Ministry of Health and Welfare and the National Research Foundation of Korea.
2025.07.08 View 270 -
 KAIST Presents a Breakthrough in Overcoming Drug Resistance in Cancer – Hope for Treating Intractable Diseases like Diabetes
<(From the left) Prof. Hyun Uk Kim, Ph.D candiate Hae Deok Jung, Ph.D candidate Jina Lim, Prof.Yoosik Kim from the Department of Chemical and Biomolecular Engineering>
One of the biggest obstacles in cancer treatment is drug resistance in cancer cells. Conventional efforts have focused on identifying new drug targets to eliminate these resistant cells, but such approaches can often lead to even stronger resistance. Now, researchers at KAIST have developed a computational framework to predict key metabolic genes that can re-sensitize resistant cancer cells to treatment. This technique holds promise not only for a variety of cancer therapies but also for treating metabolic diseases such as diabetes.
On the 7th of July, KAIST (President Kwang Hyung Lee) announced that a research team led by Professors Hyun Uk Kim and Yoosik Kim from the Department of Chemical and Biomolecular Engineering had developed a computational framework that predicts metabolic gene targets to re-sensitize drug-resistant breast cancer cells. This was achieved using a metabolic network model capable of simulating human metabolism.
Focusing on metabolic alterations—key characteristics in the formation of drug resistance—the researchers developed a metabolism-based approach to identify gene targets that could enhance drug responsiveness by regulating the metabolism of drug-resistant breast cancer cells.
< Computational framework that can identify metabolic gene targets to revert the metabolic state of the drug-resistant cells to that of the drug-sensitive parental cells>
The team first constructed cell-specific metabolic network models by integrating proteomic data obtained from two different types of drug-resistant MCF7 breast cancer cell lines: one resistant to doxorubicin and the other to paclitaxel. They then performed gene knockout simulations* on all of the metabolic genes and analyzed the results.
*Gene knockout simulation: A computational method to predict changes in a biological network by virtually removing specific genes.
As a result, they discovered that suppressing certain genes could make previously resistant cancer cells responsive to anticancer drugs again. Specifically, they identified GOT1 as a target in doxorubicin-resistant cells, GPI in paclitaxel-resistant cells, and SLC1A5 as a common target for both drugs.
The predictions were experimentally validated by suppressing proteins encoded by these genes, which led to the re-sensitization of the drug-resistant cancer cells.
Furthermore, consistent re-sensitization effects were also observed when the same proteins were inhibited in other types of breast cancer cells that had developed resistance to the same drugs.
Professor Yoosik Kim remarked, “Cellular metabolism plays a crucial role in various intractable diseases including infectious and degenerative conditions. This new technology, which predicts metabolic regulation switches, can serve as a foundational tool not only for treating drug-resistant breast cancer but also for a wide range of diseases that currently lack effective therapies.”
Professor Hyun Uk Kim, who led the study, emphasized, “The significance of this research lies in our ability to accurately predict key metabolic genes that can make resistant cancer cells responsive to treatment again—using only computer simulations and minimal experimental data. This framework can be widely applied to discover new therapeutic targets in various cancers and metabolic diseases.”
The study, in which Ph.D. candidates JinA Lim and Hae Deok Jung from KAIST participated as co-first authors, was published online on June 25 in Proceedings of the National Academy of Sciences (PNAS), a leading multidisciplinary journal that covers top-tier research in life sciences, physics, engineering, and social sciences.
※ Title: Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets ※ DOI: https://doi.org/10.1073/pnas.2425384122 ※ Authors: JinA Lim (KAIST, co-first author), Hae Deok Jung (KAIST, co-first author), Han Suk Ryu (Seoul National University Hospital, corresponding author), Yoosik Kim (KAIST, corresponding author), Hyun Uk Kim (KAIST, corresponding author), and five others.
This research was supported by the Ministry of Science and ICT through the National Research Foundation of Korea, and the Electronics and Telecommunications Research Institute (ETRI).
2025.07.08 View 374
KAIST Presents a Breakthrough in Overcoming Drug Resistance in Cancer – Hope for Treating Intractable Diseases like Diabetes
<(From the left) Prof. Hyun Uk Kim, Ph.D candiate Hae Deok Jung, Ph.D candidate Jina Lim, Prof.Yoosik Kim from the Department of Chemical and Biomolecular Engineering>
One of the biggest obstacles in cancer treatment is drug resistance in cancer cells. Conventional efforts have focused on identifying new drug targets to eliminate these resistant cells, but such approaches can often lead to even stronger resistance. Now, researchers at KAIST have developed a computational framework to predict key metabolic genes that can re-sensitize resistant cancer cells to treatment. This technique holds promise not only for a variety of cancer therapies but also for treating metabolic diseases such as diabetes.
On the 7th of July, KAIST (President Kwang Hyung Lee) announced that a research team led by Professors Hyun Uk Kim and Yoosik Kim from the Department of Chemical and Biomolecular Engineering had developed a computational framework that predicts metabolic gene targets to re-sensitize drug-resistant breast cancer cells. This was achieved using a metabolic network model capable of simulating human metabolism.
Focusing on metabolic alterations—key characteristics in the formation of drug resistance—the researchers developed a metabolism-based approach to identify gene targets that could enhance drug responsiveness by regulating the metabolism of drug-resistant breast cancer cells.
< Computational framework that can identify metabolic gene targets to revert the metabolic state of the drug-resistant cells to that of the drug-sensitive parental cells>
The team first constructed cell-specific metabolic network models by integrating proteomic data obtained from two different types of drug-resistant MCF7 breast cancer cell lines: one resistant to doxorubicin and the other to paclitaxel. They then performed gene knockout simulations* on all of the metabolic genes and analyzed the results.
*Gene knockout simulation: A computational method to predict changes in a biological network by virtually removing specific genes.
As a result, they discovered that suppressing certain genes could make previously resistant cancer cells responsive to anticancer drugs again. Specifically, they identified GOT1 as a target in doxorubicin-resistant cells, GPI in paclitaxel-resistant cells, and SLC1A5 as a common target for both drugs.
The predictions were experimentally validated by suppressing proteins encoded by these genes, which led to the re-sensitization of the drug-resistant cancer cells.
Furthermore, consistent re-sensitization effects were also observed when the same proteins were inhibited in other types of breast cancer cells that had developed resistance to the same drugs.
Professor Yoosik Kim remarked, “Cellular metabolism plays a crucial role in various intractable diseases including infectious and degenerative conditions. This new technology, which predicts metabolic regulation switches, can serve as a foundational tool not only for treating drug-resistant breast cancer but also for a wide range of diseases that currently lack effective therapies.”
Professor Hyun Uk Kim, who led the study, emphasized, “The significance of this research lies in our ability to accurately predict key metabolic genes that can make resistant cancer cells responsive to treatment again—using only computer simulations and minimal experimental data. This framework can be widely applied to discover new therapeutic targets in various cancers and metabolic diseases.”
The study, in which Ph.D. candidates JinA Lim and Hae Deok Jung from KAIST participated as co-first authors, was published online on June 25 in Proceedings of the National Academy of Sciences (PNAS), a leading multidisciplinary journal that covers top-tier research in life sciences, physics, engineering, and social sciences.
※ Title: Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets ※ DOI: https://doi.org/10.1073/pnas.2425384122 ※ Authors: JinA Lim (KAIST, co-first author), Hae Deok Jung (KAIST, co-first author), Han Suk Ryu (Seoul National University Hospital, corresponding author), Yoosik Kim (KAIST, corresponding author), Hyun Uk Kim (KAIST, corresponding author), and five others.
This research was supported by the Ministry of Science and ICT through the National Research Foundation of Korea, and the Electronics and Telecommunications Research Institute (ETRI).
2025.07.08 View 374 -
 KAIST Enhances Immunotherapy for Difficult-to-Treat Brain Tumors with Gut Microbiota
< Photo 1.(From left) Prof. Heung Kyu Lee, Department of Biological Sciences,
and Dr. Hyeon Cheol Kim>
Advanced treatments, known as immunotherapies that activate T cells—our body's immune cells—to eliminate cancer cells, have shown limited efficacy as standalone therapies for glioblastoma, the most lethal form of brain tumor. This is due to their minimal response to glioblastoma and high resistance to treatment.
Now, a KAIST research team has now demonstrated a new therapeutic strategy that can enhance the efficacy of immunotherapy for brain tumors by utilizing gut microbes and their metabolites. This also opens up possibilities for developing microbiome-based immunotherapy supplements in the future.
KAIST (President Kwang Hyung Lee) announced on July 1 that a research team led by Professor Heung Kyu Lee of the Department of Biological Sciences discovered and demonstrated a method to significantly improve the efficiency of glioblastoma immunotherapy by focusing on changes in the gut microbial ecosystem.
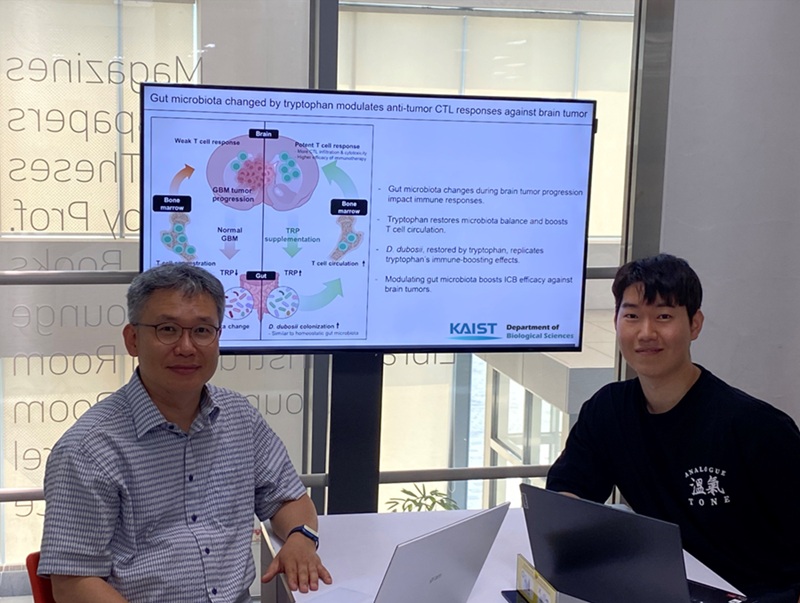
The research team noted that as glioblastoma progresses, the concentration of ‘tryptophan’, an important amino acid in the gut, sharply decreases, leading to changes in the gut microbial ecosystem. They discovered that by supplementing tryptophan to restore microbial diversity, specific beneficial strains activate CD8 T cells (a type of immune cell) and induce their infiltration into tumor tissues. Through a mouse model of glioblastoma, the research team confirmed that tryptophan supplementation enhanced the response of cancer-attacking T cells (especially CD8 T cells), leading to their increased migration to tumor sites such as lymph nodes and the brain.
In this process, they also revealed that ‘Duncaniella dubosii’, a beneficial commensal bacterium present in the gut, plays a crucial role. This bacterium helped T cells effectively redistribute within the body, and survival rates significantly improved when used in combination with immunotherapy (anti-PD-1).
Furthermore, it was demonstrated that even when this commensal bacterium was administered alone to germ-free mice (mice without any commensal microbes), the survival rate for glioblastoma increased. This is because the bacterium utilizes tryptophan to regulate the gut environment, and the metabolites produced in this process strengthen the ability of CD8 T cells to attack cancer cells.
Professor Heung Kyu Lee explained, "This research is a meaningful achievement, showing that even in intractable brain tumors where immune checkpoint inhibitors had no effect, a combined strategy utilizing gut microbes can significantly enhance treatment response."
Dr. Hyeon Cheol Kim of KAIST (currently a postdoctoral researcher at the Institute for Biological Sciences) participated as the first author. The research findings were published online in Cell Reports, an international journal in the life sciences, on June 26.
This research was conducted as part of the Basic Research Program and Bio & Medical Technology Development Program supported by the Ministry of Science and ICT and the National Research Foundation of Korea.
※Paper Title: Gut microbiota dysbiosis induced by brain tumor modulates the efficacy of immunotherapy
※DOI: https://doi.org/10.1016/j.celrep.2025.115825
2025.07.02 View 842
KAIST Enhances Immunotherapy for Difficult-to-Treat Brain Tumors with Gut Microbiota
< Photo 1.(From left) Prof. Heung Kyu Lee, Department of Biological Sciences,
and Dr. Hyeon Cheol Kim>
Advanced treatments, known as immunotherapies that activate T cells—our body's immune cells—to eliminate cancer cells, have shown limited efficacy as standalone therapies for glioblastoma, the most lethal form of brain tumor. This is due to their minimal response to glioblastoma and high resistance to treatment.
Now, a KAIST research team has now demonstrated a new therapeutic strategy that can enhance the efficacy of immunotherapy for brain tumors by utilizing gut microbes and their metabolites. This also opens up possibilities for developing microbiome-based immunotherapy supplements in the future.
KAIST (President Kwang Hyung Lee) announced on July 1 that a research team led by Professor Heung Kyu Lee of the Department of Biological Sciences discovered and demonstrated a method to significantly improve the efficiency of glioblastoma immunotherapy by focusing on changes in the gut microbial ecosystem.
The research team noted that as glioblastoma progresses, the concentration of ‘tryptophan’, an important amino acid in the gut, sharply decreases, leading to changes in the gut microbial ecosystem. They discovered that by supplementing tryptophan to restore microbial diversity, specific beneficial strains activate CD8 T cells (a type of immune cell) and induce their infiltration into tumor tissues. Through a mouse model of glioblastoma, the research team confirmed that tryptophan supplementation enhanced the response of cancer-attacking T cells (especially CD8 T cells), leading to their increased migration to tumor sites such as lymph nodes and the brain.
In this process, they also revealed that ‘Duncaniella dubosii’, a beneficial commensal bacterium present in the gut, plays a crucial role. This bacterium helped T cells effectively redistribute within the body, and survival rates significantly improved when used in combination with immunotherapy (anti-PD-1).
Furthermore, it was demonstrated that even when this commensal bacterium was administered alone to germ-free mice (mice without any commensal microbes), the survival rate for glioblastoma increased. This is because the bacterium utilizes tryptophan to regulate the gut environment, and the metabolites produced in this process strengthen the ability of CD8 T cells to attack cancer cells.
Professor Heung Kyu Lee explained, "This research is a meaningful achievement, showing that even in intractable brain tumors where immune checkpoint inhibitors had no effect, a combined strategy utilizing gut microbes can significantly enhance treatment response."
Dr. Hyeon Cheol Kim of KAIST (currently a postdoctoral researcher at the Institute for Biological Sciences) participated as the first author. The research findings were published online in Cell Reports, an international journal in the life sciences, on June 26.
This research was conducted as part of the Basic Research Program and Bio & Medical Technology Development Program supported by the Ministry of Science and ICT and the National Research Foundation of Korea.
※Paper Title: Gut microbiota dysbiosis induced by brain tumor modulates the efficacy of immunotherapy
※DOI: https://doi.org/10.1016/j.celrep.2025.115825
2025.07.02 View 842 -
 KAIST develops technology for selective RNA modification in living cells and animals
· A team led by Professor Won Do Heo from the Department of Biological Sciences, KAIST, has developed a pioneering technology that selectively acetylates specific RNA molecules in living cells and tissues.
· The platform uses RNA-targeting CRISPR tools in combination with RNA-modifying enzymes to chemically modify only the intended RNA.
· The method opens new possibilities for gene therapy by enabling precise control of disease-related RNA without affecting the rest of the transcriptome.
< Photo 1. (From left) Professor Won Do Heo and Jihwan Yu, a Ph.D. Candidate of the Department of Biological Sciences >
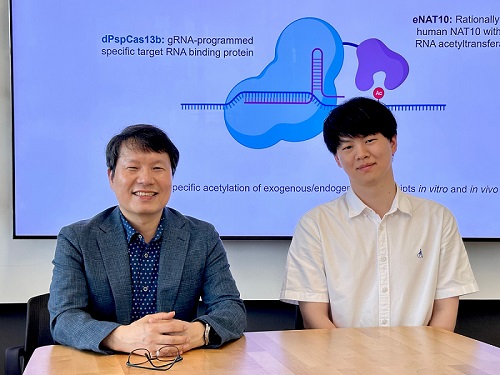
CRISPR-Cas13, a powerful RNA-targeting technology is gaining increasing attention as a next-generation gene therapy platform due to its precision and reduced side effects. Utilizing this system, researchers at KAIST have now developed the world’s first technology capable of selectively acetylating (chemically modifying) specific RNA molecules among countless transcripts within living cells. This breakthrough enables precise, programmable control of RNA function and is expected to open new avenues in RNA-based therapeutic development.
KAIST (President Kwang Hyung Lee) announced that a research team led by Professor Won Do Heo in the Department of Biological Sciences has recently developed a groundbreaking technology capable of selectively acetylating specific RNA molecules within the human body using the CRISPR-Cas13 system—an RNA-targeting platform gaining increasing attention in the fields of gene regulation and RNA-based therapeutics.
RNA molecules can undergo chemical modifications—the addition of specific chemical groups—which alter their function and behavior without changing the underlying nucleotide sequence. However, some of these modifications, a critical layer of post-transcriptional gene regulation, remain poorly understood. Among them, N4-acetylcytidine (ac4C) has been particularly enigmatic, with ongoing debate about its existence and function in human messenger RNA (mRNA), the RNA that encodes proteins.
To address this gap, the KAIST research team developed a targeted RNA acetylation system, named dCas13-eNAT10. This platform combines a catalytically inactive Cas13 enzyme (dCas13) that guides the system to specific RNA targets, with a hyperactive variant of the NAT10 enzyme (eNAT10), which performs RNA acetylation. This approach enables precise acetylation of only the desired RNA molecules among the vast pool of transcripts within the cell.
< Figure 1. Development of hyperactive variant eNAT10 through NAT10 protein engineering. By engineering the NAT10 protein, which performs RNA acetylation in human cells, based on its domain and structure, eNAT10 was developed, showing approximately a 3-fold increase in RNA acetylation activity compared to the wild-type enzyme. >
Using this system, the researchers demonstrated that guide RNAs could direct the dCas13-eNAT10 complex to acetylate specific RNA targets, and acetylation significantly increased protein expression from the modified mRNA. Moreover, the study revealed, for the first time, that RNA acetylation plays a role in intracellular RNA localization, facilitating the export of RNA from the nucleus to the cytoplasm—a critical step in gene expression regulation.
To validate its therapeutic potential, the team successfully delivered the targeted RNA acetylation system into the livers of live mice using adeno-associated virus (AAV), a commonly used gene therapy vector. This marks the first demonstration of in vivo RNA modification, extending the applicability of RNA chemical modification tools from cell culture models to living organisms.
< Figure 2. Acetylation of various RNA in cells using dCas13-eNAT10 fusion protein. Utilizing the CRISPR-Cas13 system, which can precisely target specific RNA through guide RNA, a dCas13-eNAT10 fusion protein was created, demonstrating its ability to specifically acetylate various endogenous RNA at different locations within cells. >
Professor Won Do Heo, who previously developed COVID-19 treatment technology using RNA gene scissors and technology to activate RNA gene scissors with light, stated, "Existing RNA chemical modification research faced difficulties in controlling specificity, temporality, and spatiality. However, this new technology allows selective acetylation of desired RNA, opening the door for accurate and detailed research into the functions of RNA acetylation." He added, "The RNA chemical modification technology developed in this study can be widely used as an RNA-based therapeutic agent and a tool for regulating RNA functions in living organisms in the future."
< Figure 3. In vivo delivery of targeted RNA acetylation system. The targeted RNA acetylation system was encoded in an AAV vector, commonly used in gene therapy, and delivered intravenously to adult mice, showing that target RNA in liver tissue was specifically acetylated according to the guide RNA. >
This research, with Ph.D. candidate Jihwan Yu from the Department of Biological Sciences at KAIST as the first author, was published in the journal Nature Chemical Biology on June 2, 2025. (Title: Programmable RNA acetylation with CRISPR-Cas13, Impact factor: 12.9, DOI: https://doi.org/10.1038/s41589-025-01922-3)
This research was supported by the Samsung Future Technology Foundation and the Bio & Medical Technology Development Program of the National Research Foundation of Korea.
2025.06.10 View 1918
KAIST develops technology for selective RNA modification in living cells and animals
· A team led by Professor Won Do Heo from the Department of Biological Sciences, KAIST, has developed a pioneering technology that selectively acetylates specific RNA molecules in living cells and tissues.
· The platform uses RNA-targeting CRISPR tools in combination with RNA-modifying enzymes to chemically modify only the intended RNA.
· The method opens new possibilities for gene therapy by enabling precise control of disease-related RNA without affecting the rest of the transcriptome.
< Photo 1. (From left) Professor Won Do Heo and Jihwan Yu, a Ph.D. Candidate of the Department of Biological Sciences >
CRISPR-Cas13, a powerful RNA-targeting technology is gaining increasing attention as a next-generation gene therapy platform due to its precision and reduced side effects. Utilizing this system, researchers at KAIST have now developed the world’s first technology capable of selectively acetylating (chemically modifying) specific RNA molecules among countless transcripts within living cells. This breakthrough enables precise, programmable control of RNA function and is expected to open new avenues in RNA-based therapeutic development.
KAIST (President Kwang Hyung Lee) announced that a research team led by Professor Won Do Heo in the Department of Biological Sciences has recently developed a groundbreaking technology capable of selectively acetylating specific RNA molecules within the human body using the CRISPR-Cas13 system—an RNA-targeting platform gaining increasing attention in the fields of gene regulation and RNA-based therapeutics.
RNA molecules can undergo chemical modifications—the addition of specific chemical groups—which alter their function and behavior without changing the underlying nucleotide sequence. However, some of these modifications, a critical layer of post-transcriptional gene regulation, remain poorly understood. Among them, N4-acetylcytidine (ac4C) has been particularly enigmatic, with ongoing debate about its existence and function in human messenger RNA (mRNA), the RNA that encodes proteins.
To address this gap, the KAIST research team developed a targeted RNA acetylation system, named dCas13-eNAT10. This platform combines a catalytically inactive Cas13 enzyme (dCas13) that guides the system to specific RNA targets, with a hyperactive variant of the NAT10 enzyme (eNAT10), which performs RNA acetylation. This approach enables precise acetylation of only the desired RNA molecules among the vast pool of transcripts within the cell.
< Figure 1. Development of hyperactive variant eNAT10 through NAT10 protein engineering. By engineering the NAT10 protein, which performs RNA acetylation in human cells, based on its domain and structure, eNAT10 was developed, showing approximately a 3-fold increase in RNA acetylation activity compared to the wild-type enzyme. >
Using this system, the researchers demonstrated that guide RNAs could direct the dCas13-eNAT10 complex to acetylate specific RNA targets, and acetylation significantly increased protein expression from the modified mRNA. Moreover, the study revealed, for the first time, that RNA acetylation plays a role in intracellular RNA localization, facilitating the export of RNA from the nucleus to the cytoplasm—a critical step in gene expression regulation.
To validate its therapeutic potential, the team successfully delivered the targeted RNA acetylation system into the livers of live mice using adeno-associated virus (AAV), a commonly used gene therapy vector. This marks the first demonstration of in vivo RNA modification, extending the applicability of RNA chemical modification tools from cell culture models to living organisms.
< Figure 2. Acetylation of various RNA in cells using dCas13-eNAT10 fusion protein. Utilizing the CRISPR-Cas13 system, which can precisely target specific RNA through guide RNA, a dCas13-eNAT10 fusion protein was created, demonstrating its ability to specifically acetylate various endogenous RNA at different locations within cells. >
Professor Won Do Heo, who previously developed COVID-19 treatment technology using RNA gene scissors and technology to activate RNA gene scissors with light, stated, "Existing RNA chemical modification research faced difficulties in controlling specificity, temporality, and spatiality. However, this new technology allows selective acetylation of desired RNA, opening the door for accurate and detailed research into the functions of RNA acetylation." He added, "The RNA chemical modification technology developed in this study can be widely used as an RNA-based therapeutic agent and a tool for regulating RNA functions in living organisms in the future."
< Figure 3. In vivo delivery of targeted RNA acetylation system. The targeted RNA acetylation system was encoded in an AAV vector, commonly used in gene therapy, and delivered intravenously to adult mice, showing that target RNA in liver tissue was specifically acetylated according to the guide RNA. >
This research, with Ph.D. candidate Jihwan Yu from the Department of Biological Sciences at KAIST as the first author, was published in the journal Nature Chemical Biology on June 2, 2025. (Title: Programmable RNA acetylation with CRISPR-Cas13, Impact factor: 12.9, DOI: https://doi.org/10.1038/s41589-025-01922-3)
This research was supported by the Samsung Future Technology Foundation and the Bio & Medical Technology Development Program of the National Research Foundation of Korea.
2025.06.10 View 1918 -
 KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 3042
KAIST-UIUC researchers develop a treatment platform to disable the ‘biofilm’ shield of superbugs
< (From left) Ph.D. Candidate Joo Hun Lee (co-author), Professor Hyunjoon Kong (co-corresponding author) and Postdoctoral Researcher Yujin Ahn (co-first author) from the Department of Chemical and Biomolecular Engineering of the University of Illinois at Urbana-Champaign and Ju Yeon Chung (co-first author) from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung (co-corresponding author) from the Department of Biological Sciences of KAIST >
A major cause of hospital-acquired infections, the super bacteria Methicillin-resistant Staphylococcus aureus (MRSA), not only exhibits strong resistance to existing antibiotics but also forms a dense biofilm that blocks the effects of external treatments. To meet this challenge, KAIST researchers, in collaboration with an international team, successfully developed a platform that utilizes microbubbles to deliver gene-targeted nanoparticles capable of break ing down the biofilms, offering an innovative solution for treating infections resistant to conventional antibiotics.
KAIST (represented by President Kwang Hyung Lee) announced on May 29 that a research team led by Professor Hyun Jung Chung from the Department of Biological Sciences, in collaboration with Professor Hyunjoon Kong's team at the University of Illinois, has developed a microbubble-based nano-gene delivery platform (BTN MB) that precisely delivers gene suppressors into bacteria to effectively remove biofilms formed by MRSA.
The research team first designed short DNA oligonucleotides that simultaneously suppress three major MRSA genes, related to—biofilm formation (icaA), cell division (ftsZ), and antibiotic resistance (mecA)—and engineered nanoparticles (BTN) to effectively deliver them into the bacteria.
< Figure 1. Effective biofilm treatment using biofilm-targeting nanoparticles controlled by microbubbler system. Schematic illustration of BTN delivery with microbubbles (MB), enabling effective permeation of ASOs targeting bacterial genes within biofilms infecting skin wounds. Gene silencing of targets involved in biofilm formation, bacterial proliferation, and antibiotic resistance leads to effective biofilm removal and antibacterial efficacy in vivo. >
In addition, microbubbles (MB) were used to increase the permeability of the microbial membrane, specifically the biofilm formed by MRSA. By combining these two technologies, the team implemented a dual-strike strategy that fundamentally blocks bacterial growth and prevents resistance acquisition.
This treatment system operates in two stages. First, the MBs induce pressure changes within the bacterial biofilm, allowing the BTNs to penetrate. Then, the BTNs slip through the gaps in the biofilm and enter the bacteria, delivering the gene suppressors precisely. This leads to gene regulation within MRSA, simultaneously blocking biofilm regeneration, cell proliferation, and antibiotic resistance expression.
In experiments conducted in a porcine skin model and a mouse wound model infected with MRSA biofilm, the BTN MB treatment group showed a significant reduction in biofilm thickness, as well as remarkable decreases in bacterial count and inflammatory responses.
< Figure 2. (a) Schematic illustration on the evaluation of treatment efficacy of BTN-MB gene therapy. (b) Reduction in MRSA biofilm mass via simultaneous inhibition of multiple genes. (c, d) Antibacterial efficacy of BTN-MB over time in a porcine skin infection biofilm model. (e) Schematic of the experimental setup to verify antibacterial efficacy in a mouse skin wound infection model. (f) Wound healing effects in mice. (g) Antibacterial effects at the wound site. (h) Histological analysis results. >
These results are difficult to achieve with conventional antibiotic monotherapy and demonstrate the potential for treating a wide range of resistant bacterial infections.
Professor Hyun Jung Chung of KAIST, who led the research, stated, “This study presents a new therapeutic solution that combines nanotechnology, gene suppression, and physical delivery strategies to address superbug infections that existing antibiotics cannot resolve. We will continue our research with the aim of expanding its application to systemic infections and various other infectious diseases.”
< (From left) Ju Yeon Chung from the Integrated Master's and Doctoral Program, and Professor Hyun Jung Chung from the Department of Biological Sciences >
The study was co-first authored by Ju Yeon Chung, a graduate student in the Department of Biological Sciences at KAIST, and Dr. Yujin Ahn from the University of Illinois. The study was published online on May 19 in the journal, Advanced Functional Materials.
※ Paper Title: Microbubble-Controlled Delivery of Biofilm-Targeting Nanoparticles to Treat MRSA Infection ※ DOI: https://doi.org/10.1002/adfm.202508291
This study was supported by the National Research Foundation and the Ministry of Health and Welfare, Republic of Korea; and the National Science Foundation and National Institutes of Health, USA.
2025.05.29 View 3042 -
 KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 4660
KAIST to Develop a Korean-style ChatGPT Platform Specifically Geared Toward Medical Diagnosis and Drug Discovery
On May 23rd, KAIST (President Kwang-Hyung Lee) announced that its Digital Bio-Health AI Research Center (Director: Professor JongChul Ye of KAIST Kim Jaechul Graduate School of AI) has been selected for the Ministry of Science and ICT's 'AI Top-Tier Young Researcher Support Program (AI Star Fellowship Project).' With a total investment of ₩11.5 billion from May 2025 to December 2030, the center will embark on the full-scale development of AI technology and a platform capable of independently inferring and determining the kinds of diseases, and discovering new drugs.
< Photo. On May 20th, a kick-off meeting for the AI Star Fellowship Project was held at KAIST Kim Jaechul Graduate School of AI’s Yangjae Research Center with the KAIST research team and participating organizations of Samsung Medical Center, NAVER Cloud, and HITS. [From left to right in the front row] Professor Jaegul Joo (KAIST), Professor Yoonjae Choi (KAIST), Professor Woo Youn Kim (KAIST/HITS), Professor JongChul Ye (KAIST), Professor Sungsoo Ahn (KAIST), Dr. Haanju Yoo (NAVER Cloud), Yoonho Lee (KAIST), HyeYoon Moon (Samsung Medical Center), Dr. Su Min Kim (Samsung Medical Center) >
This project aims to foster an innovative AI research ecosystem centered on young researchers and develop an inferential AI agent that can utilize and automatically expand specialized knowledge systems in the bio and medical fields.
Professor JongChul Ye of the Kim Jaechul Graduate School of AI will serve as the lead researcher, with young researchers from KAIST including Professors Yoonjae Choi, Kimin Lee, Sungsoo Ahn, and Chanyoung Park, along with mid-career researchers like Professors Jaegul Joo and Woo Youn Kim, jointly undertaking the project. They will collaborate with various laboratories within KAIST to conduct comprehensive research covering the entire cycle from the theoretical foundations of AI inference to its practical application.
Specifically, the main goals include: - Building high-performance inference models that integrate diverse medical knowledge systems to enhance the precision and reliability of diagnosis and treatment. - Developing a convergence inference platform that efficiently combines symbol-based inference with neural network models. - Securing AI technology for new drug development and biomarker discovery based on 'cell ontology.'
Furthermore, through close collaboration with industry and medical institutions such as Samsung Medical Center, NAVER Cloud, and HITS Co., Ltd., the project aims to achieve: - Clinical diagnostic AI utilizing medical knowledge systems. - AI-based molecular target exploration for new drug development. - Commercialization of an extendible AI inference platform.
Professor JongChul Ye, Director of KAIST's Digital Bio-Health AI Research Center, stated, "At a time when competition in AI inference model development is intensifying, it is a great honor for KAIST to lead the development of AI technology specialized in the bio and medical fields with world-class young researchers." He added, "We will do our best to ensure that the participating young researchers reach a world-leading level in terms of research achievements after the completion of this seven-year project starting in 2025."
The AI Star Fellowship is a newly established program where post-doctoral researchers and faculty members within seven years of appointment participate as project leaders (PLs) to independently lead research. Multiple laboratories within a university and demand-side companies form a consortium to operate the program.
Through this initiative, KAIST plans to nurture bio-medical convergence AI talent and simultaneously promote the commercialization of core technologies in collaboration with Samsung Medical Center, NAVER Cloud, and HITS.
2025.05.26 View 4660 -
 KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 6467
KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 6467 -
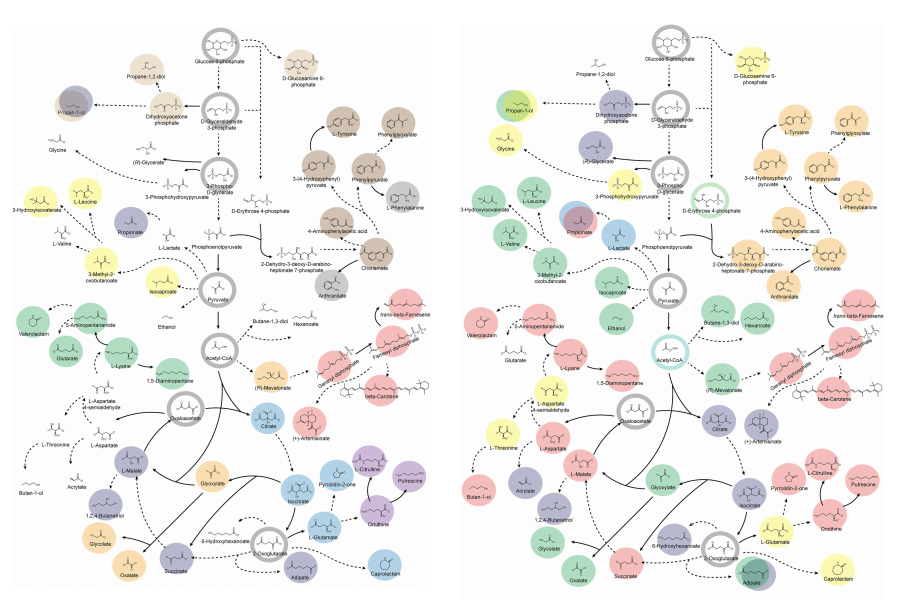 KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 4741
KAIST provides a comprehensive resource on microbial cell factories for sustainable chemical production
In silico analysis of five industrial microorganisms identifies optimal strains and metabolic engineering strategies for producing 235 valuable chemicals
Climate change and the depletion of fossil fuels have raised the global need for sustainable chemical production. In response to these environmental challenges, microbial cell factories are gaining attention as eco-friendly platforms for producing chemicals using renewable resources, while metabolic engineering technologies to enhance these cell factories are becoming crucial tools for maximizing production efficiency. However, difficulties in selecting suitable microbial strains and optimizing complex metabolic pathways continue to pose significant obstacles to practical industrial applications.
KAIST (President Kwang-Hyung Lee) announced on 27th of March that Distinguished Professor Sang Yup Lee’s research team in the Department of Chemical and Biomolecular Engineering comprehensively evaluated the production capabilities of various industrial microbial cell factories using in silico simulations and, based on these findings, identified the most suitable microbial strains for producing specific chemicals as well as optimal metabolic engineering strategies.
Previously, researchers attempted to determine the best strains and efficient metabolic engineering strategies among numerous microbial candidates through extensive biological experiments and meticulous verification processes. However, this approach required substantial time and costs. Recently, the introduction of genome-scale metabolic models (GEMs), which reconstruct the metabolic networks within an organism based on its entire genome information, has enabled systematic analysis of metabolic fluxes via computer simulations. This development offers a new way to overcome limitations of conventional experimental approaches, revolutionizing both strain selection and metabolic pathway design.
Accordingly, Professor Lee’s team at the Department of Chemical and Biomolecular Engineering, KAIST, evaluated the production capabilities of five representative industrial microorganisms—Escherichia coli, Saccharomyces cerevisiae, Bacillus subtilis, Corynebacterium glutamicum, and Pseudomonas putida—for 235 bio-based chemicals. Using GEMs, the researchers calculated both the maximum theoretical yields and the maximum achievable yields under industrial conditions for each chemical, thereby establishing criteria to identify the most suitable strains for each target compound.
< Figure 1. Outline of the strategy for improving microbial cell factories using a genome-scale metabolic model (GEM) >
The team specifically proposed strategies such as introducing heterologous enzyme reactions derived from other organisms and exchanging cofactors used by microbes to expand metabolic pathways. These strategies were shown to increase yields beyond the innate metabolic capacities of the microorganisms, resulting in higher production of industrially important chemicals such as mevalonic acid, propanol, fatty acids, and isoprenoids.
Moreover, by applying a computational approach to analyze metabolic fluxes in silico, the researchers suggested strategies for improving microbial strains to maximize the production of various chemicals. They quantitatively identified the relationships between specific enzyme reactions and target chemical production, as well as the relationships between enzymes and metabolites, determining which enzyme reactions should be up- or down-regulated. Through this, the team presented strategies not only to achieve high theoretical yields but also to maximize actual production capacities.
< Figure 2. Comparison of production routes and maximum yields of useful chemicals using representative industrial microorganisms >
Dr. Gi Bae Kim, the first author of this paper from the KAIST BioProcess Engineering Research Center, explained, “By introducing metabolic pathways derived from other organisms and exchanging cofactors, it is possible to design new microbial cell factories that surpass existing limitations. The strategies presented in this study will play a pivotal role in making microbial-based production processes more economical and efficient.” In addition, Distinguished Professor Sang Yup Lee noted, “This research serves as a key resource in the field of systems metabolic engineering, reducing difficulties in strain selection and pathway design, and enabling more efficient development of microbial cell factories. We expect it to greatly contribute to the future development of technologies for producing various eco-friendly chemicals, such as biofuels, bioplastics, and functional food materials.”
This research was conducted with the support from the Development of platform technologies of microbial cell factories for the next-generation biorefineries project and Development of advanced synthetic biology source technologies for leading the biomanufacturing industry project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) from National Research Foundation supported by the Korean Ministry of Science and ICT.
2025.03.27 View 4741 -
 KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 6048
KAIST Develops Eco-Friendly, Nylon-Like Plastic Using Microorganisms
Poly(ester amide) amide is a next-generation material that combines the advantages of PET (polyester) and nylon (polyamide), two widely used plastics. However, it could only be produced from fossil fuels, which posed environmental concerns. Using microorganisms, KAIST researchers have successfully developed a new bio-based plastic to replace conventional plastic.
KAIST (represented by President Kwang Hyung Lee) announced on the 20th of March that a research team led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering has developed microbial strains through systems metabolic engineering to produce various eco-friendly, bio-based poly(ester amide)s. The team collaborated with researchers from the Korea Research Institute of Chemical Technology (KRICT, President Young-Kook Lee) to analyze and confirm the properties of the resulting plastic.
Professor Sang Yup Lee’s research team designed new metabolic pathways that do not naturally exist in microorganisms, and developed a platform microbial strain capable of producing nine different types of poly(ester amide)s, including poly(3-hydroxybutyrate-ran-3-aminopropionate) and poly(3-hydroxybutyrate-ran-4-aminobutyrate).
Using glucose derived from abundant biomass sources such as waste wood and weeds, the team successfully produced poly(ester amide)s in an eco-friendly manner. The researchers also confirmed the potential for industrial-scale production by demonstrating high production efficiency (54.57 g/L) using fed-batch fermentation of the engineered strain.
In collaboration with researchers Haemin Jeong and Jihoon Shin from KRICT, the KAIST team analyzed the properties of the bio-based plastic and found that it exhibited characteristics similar to high-density polyethylene (HDPE). This means the new plastic is not only eco-friendly but also strong and durable enough to replace conventional plastics.
The engineered strains and strategies developed in this study are expected to be useful not only for producing various poly(ester amide)s but also for constructing metabolic pathways for the biosynthesis of other types of polymers.
Professor Sang Yup Lee stated, “This study is the first to demonstrate the possibility of producing poly(ester amide)s (plastics) through a renewable bio-based chemical process rather than relying on the petroleum-based chemical industry. We plan to further enhance the production yield and efficiency through continued research.”
The study was published online on March 17 in the international journal Nature Chemical Biology.
·Title: Biosynthesis of poly(ester amide)s in engineered Escherichia coli
·DOI: 10.1038/s41589-025-01842-2
·Authors: A total of seven authors including Tong Un Chae (KAIST, first author), So Young Choi (KAIST, second author), Da-Hee Ahn (KAIST, third author), Woo Dae Jang (KAIST, fourth author), Haemin Jeong (KRICT, fifth author), Jihoon Shin (KRICT, sixth author), and Sang Yup Lee (KAIST, corresponding author).
This research was supported by the Ministry of Science and ICT (MSIT) under the Eco-Friendly Chemical Technology Development Project as part of the "Next-Generation Biorefinery Technology Development to Lead the Bio-Chemical Industry" initiative (project led by Distinguished Professor Sang Yup Lee at KAIST).
2025.03.24 View 6048